Bromine »
PDB 1e7b-1ih6 »
1eba »
Bromine in PDB 1eba: Complex Between the Extracellular Domain of Erythropoietin (Epo) Receptor [Ebp] and An Inactive Peptide [EMP33] Contains 3,5-Dibromotyrosine in Position 4 (Denoted Dby)
Protein crystallography data
The structure of Complex Between the Extracellular Domain of Erythropoietin (Epo) Receptor [Ebp] and An Inactive Peptide [EMP33] Contains 3,5-Dibromotyrosine in Position 4 (Denoted Dby), PDB code: 1eba
was solved by
O.Livnah,
E.A.Stura,
I.A.Wilson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.70 |
| Space group | P 2 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.890, 75.009, 130.527, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20 / 29.9 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Complex Between the Extracellular Domain of Erythropoietin (Epo) Receptor [Ebp] and An Inactive Peptide [EMP33] Contains 3,5-Dibromotyrosine in Position 4 (Denoted Dby)
(pdb code 1eba). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 4 binding sites of Bromine where determined in the Complex Between the Extracellular Domain of Erythropoietin (Epo) Receptor [Ebp] and An Inactive Peptide [EMP33] Contains 3,5-Dibromotyrosine in Position 4 (Denoted Dby), PDB code: 1eba:
Jump to Bromine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Bromine where determined in the Complex Between the Extracellular Domain of Erythropoietin (Epo) Receptor [Ebp] and An Inactive Peptide [EMP33] Contains 3,5-Dibromotyrosine in Position 4 (Denoted Dby), PDB code: 1eba:
Jump to Bromine binding site number: 1; 2; 3; 4;
Bromine binding site 1 out of 4 in 1eba
Go back to
Bromine binding site 1 out
of 4 in the Complex Between the Extracellular Domain of Erythropoietin (Epo) Receptor [Ebp] and An Inactive Peptide [EMP33] Contains 3,5-Dibromotyrosine in Position 4 (Denoted Dby)
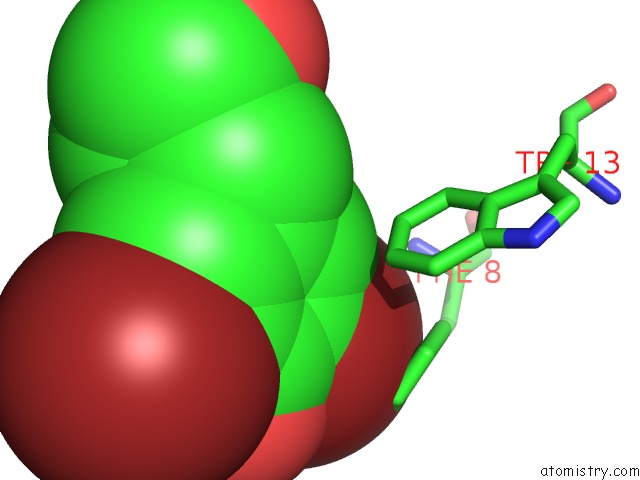
Mono view
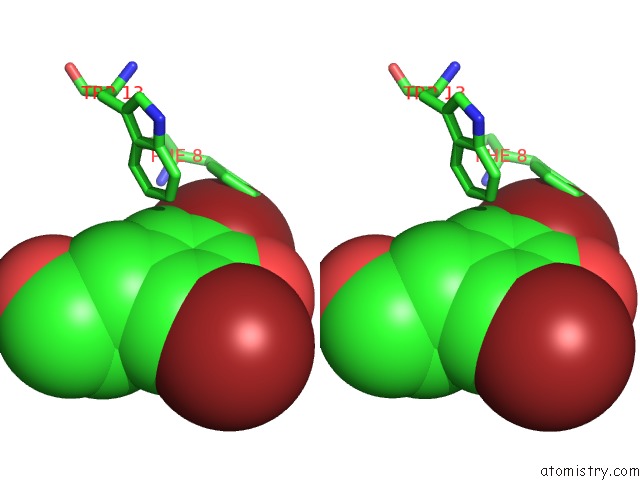
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Complex Between the Extracellular Domain of Erythropoietin (Epo) Receptor [Ebp] and An Inactive Peptide [EMP33] Contains 3,5-Dibromotyrosine in Position 4 (Denoted Dby) within 5.0Å range:
|
Bromine binding site 2 out of 4 in 1eba
Go back to
Bromine binding site 2 out
of 4 in the Complex Between the Extracellular Domain of Erythropoietin (Epo) Receptor [Ebp] and An Inactive Peptide [EMP33] Contains 3,5-Dibromotyrosine in Position 4 (Denoted Dby)
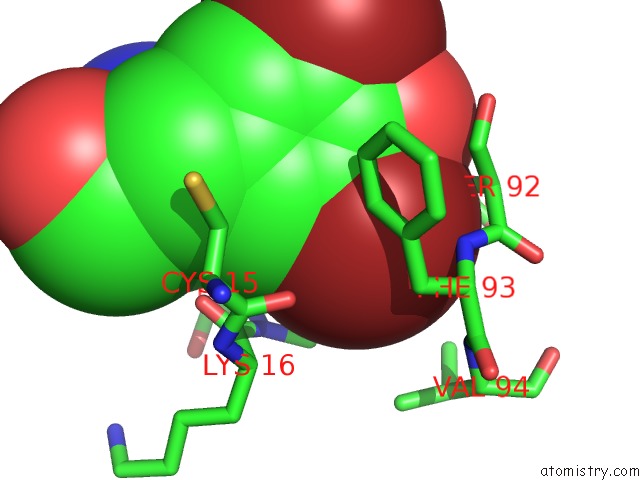
Mono view
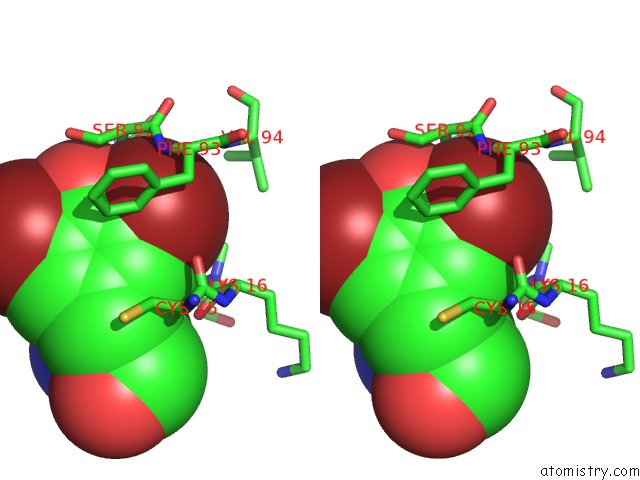
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Complex Between the Extracellular Domain of Erythropoietin (Epo) Receptor [Ebp] and An Inactive Peptide [EMP33] Contains 3,5-Dibromotyrosine in Position 4 (Denoted Dby) within 5.0Å range:
|
Bromine binding site 3 out of 4 in 1eba
Go back to
Bromine binding site 3 out
of 4 in the Complex Between the Extracellular Domain of Erythropoietin (Epo) Receptor [Ebp] and An Inactive Peptide [EMP33] Contains 3,5-Dibromotyrosine in Position 4 (Denoted Dby)
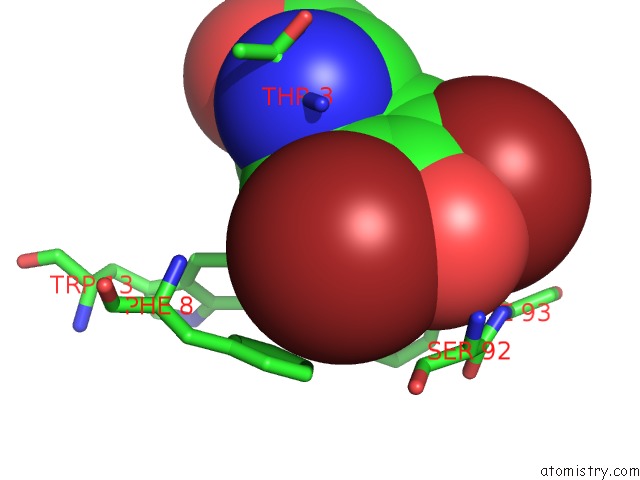
Mono view
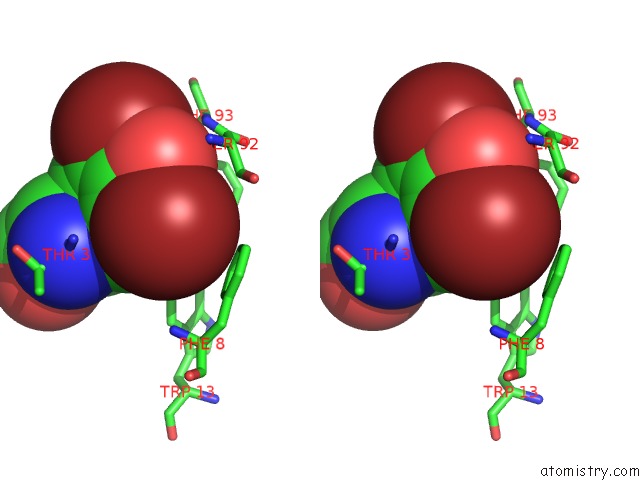
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of Complex Between the Extracellular Domain of Erythropoietin (Epo) Receptor [Ebp] and An Inactive Peptide [EMP33] Contains 3,5-Dibromotyrosine in Position 4 (Denoted Dby) within 5.0Å range:
|
Bromine binding site 4 out of 4 in 1eba
Go back to
Bromine binding site 4 out
of 4 in the Complex Between the Extracellular Domain of Erythropoietin (Epo) Receptor [Ebp] and An Inactive Peptide [EMP33] Contains 3,5-Dibromotyrosine in Position 4 (Denoted Dby)
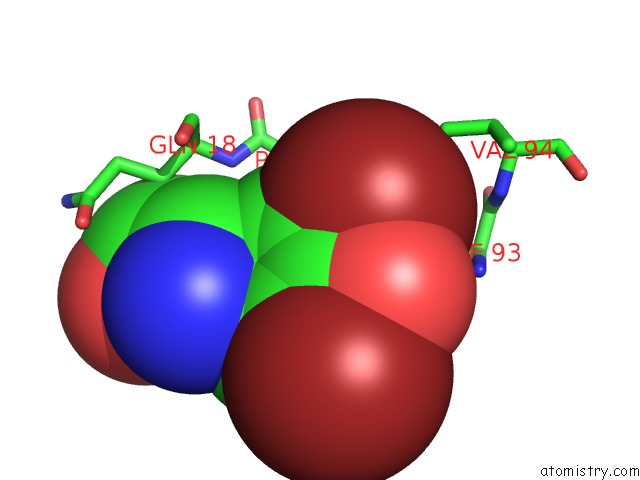
Mono view
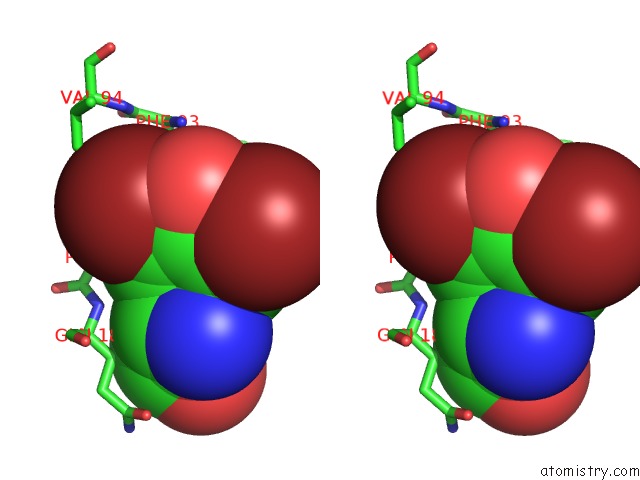
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 4 of Complex Between the Extracellular Domain of Erythropoietin (Epo) Receptor [Ebp] and An Inactive Peptide [EMP33] Contains 3,5-Dibromotyrosine in Position 4 (Denoted Dby) within 5.0Å range:
|
Reference:
O.Livnah,
D.L.Johnson,
E.A.Stura,
F.X.Farrell,
F.P.Barbone,
Y.You,
K.D.Liu,
M.A.Goldsmith,
W.He,
C.D.Krause,
S.Pestka,
L.K.Jolliffe,
I.A.Wilson.
An Antagonist Peptide-Epo Receptor Complex Suggests That Receptor Dimerization Is Not Sufficient For Activation. Nat.Struct.Biol. V. 5 993 1998.
ISSN: ISSN 1072-8368
PubMed: 9808045
DOI: 10.1038/2965
Page generated: Mon Jul 7 03:07:59 2025
ISSN: ISSN 1072-8368
PubMed: 9808045
DOI: 10.1038/2965
Last articles
F in 7F1DF in 7EW1
F in 7EX3
F in 7EU5
F in 7EVY
F in 7EVJ
F in 7EUV
F in 7EV3
F in 7EST
F in 7ET1