Bromine »
PDB 2civ-2hb1 »
2glp »
Bromine in PDB 2glp: Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) From Helicobacter Pylori Complexed with Compound 1
Protein crystallography data
The structure of Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) From Helicobacter Pylori Complexed with Compound 1, PDB code: 2glp
was solved by
L.Zhang,
W.Liu,
X.Shen,
H.Jiang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.42 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.003, 100.578, 186.472, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.9 / 23.5 |
Other elements in 2glp:
The structure of Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) From Helicobacter Pylori Complexed with Compound 1 also contains other interesting chemical elements:
| Chlorine | (Cl) | 6 atoms |
Bromine Binding Sites:
The binding sites of Bromine atom in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) From Helicobacter Pylori Complexed with Compound 1
(pdb code 2glp). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 4 binding sites of Bromine where determined in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) From Helicobacter Pylori Complexed with Compound 1, PDB code: 2glp:
Jump to Bromine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Bromine where determined in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) From Helicobacter Pylori Complexed with Compound 1, PDB code: 2glp:
Jump to Bromine binding site number: 1; 2; 3; 4;
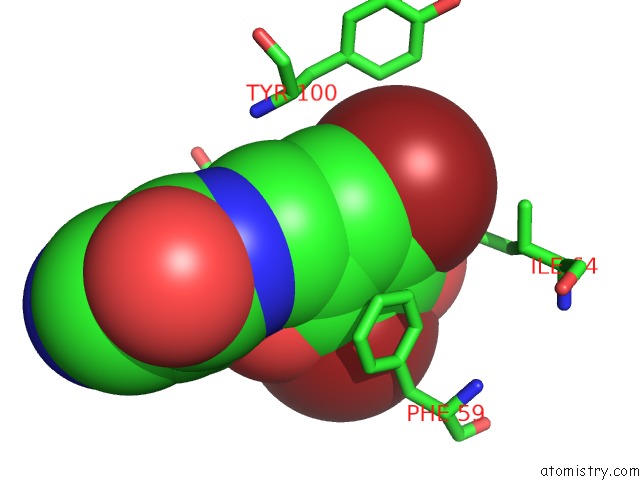
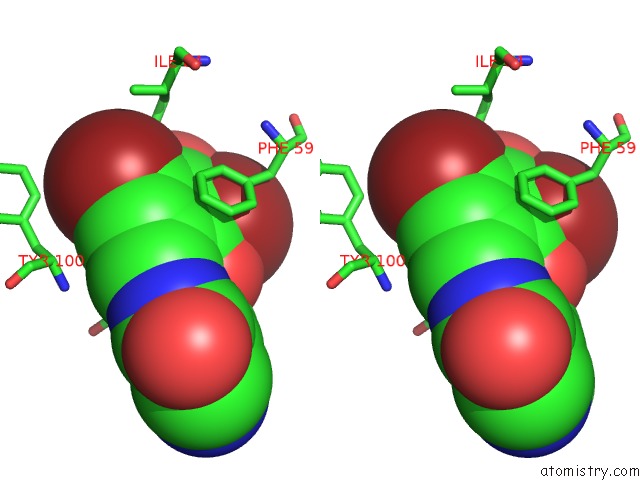
Bromine binding site 1 out of 4 in 2glp
Go back to
Bromine binding site 1 out
of 4 in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) From Helicobacter Pylori Complexed with Compound 1
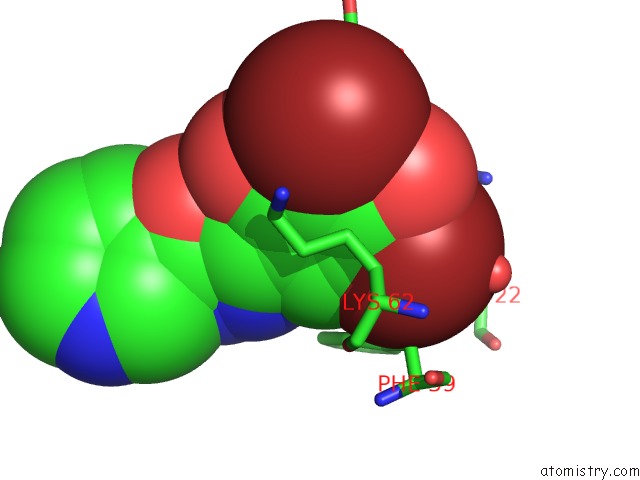
Mono view
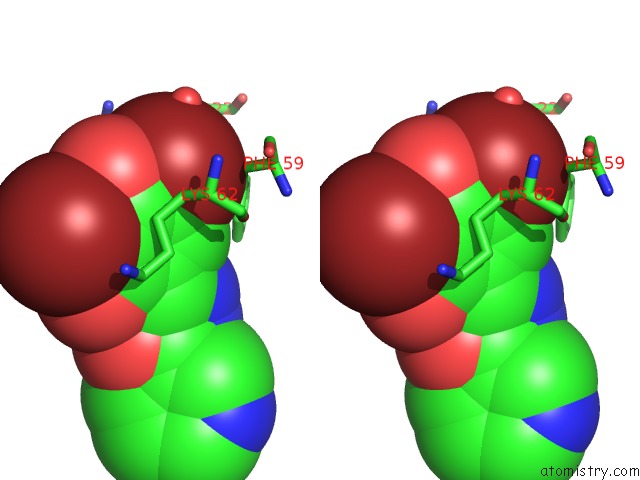
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) From Helicobacter Pylori Complexed with Compound 1 within 5.0Å range:
|
Bromine binding site 2 out of 4 in 2glp
Go back to
Bromine binding site 2 out
of 4 in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) From Helicobacter Pylori Complexed with Compound 1
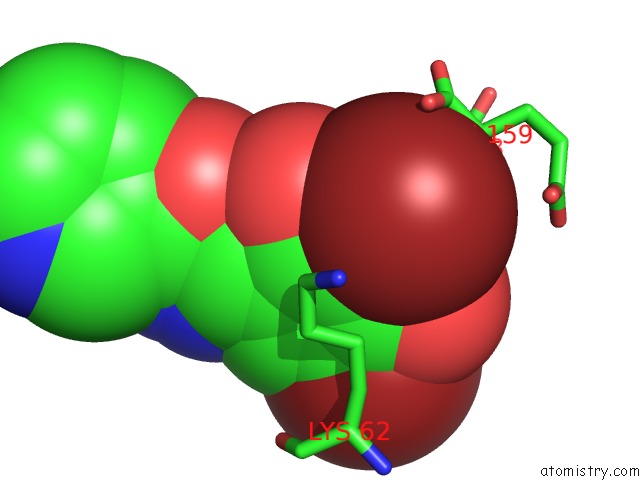
Mono view
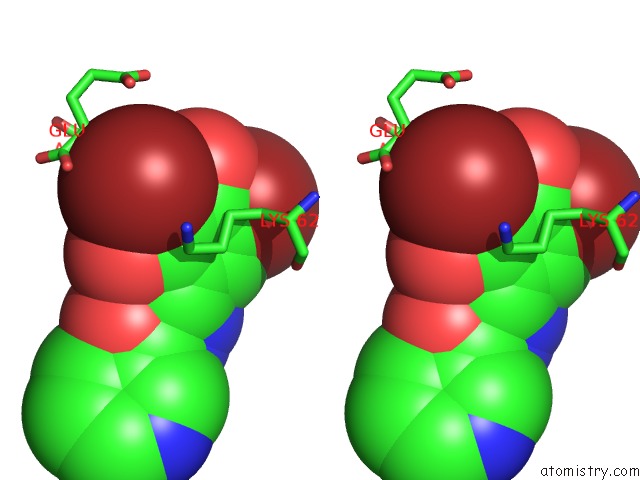
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) From Helicobacter Pylori Complexed with Compound 1 within 5.0Å range:
|
Bromine binding site 3 out of 4 in 2glp
Go back to
Bromine binding site 3 out
of 4 in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) From Helicobacter Pylori Complexed with Compound 1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) From Helicobacter Pylori Complexed with Compound 1 within 5.0Å range:
|
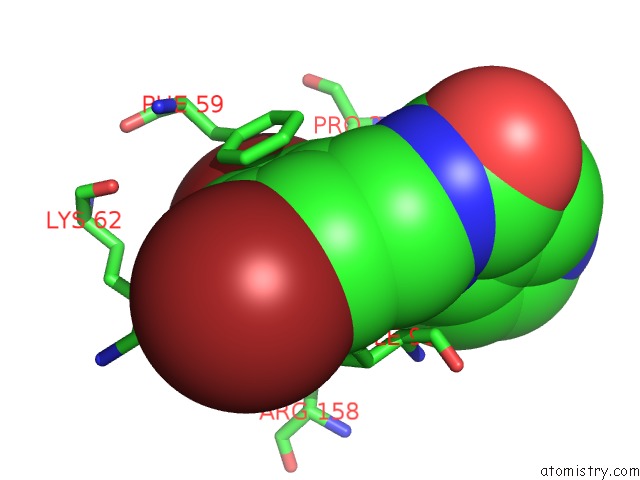
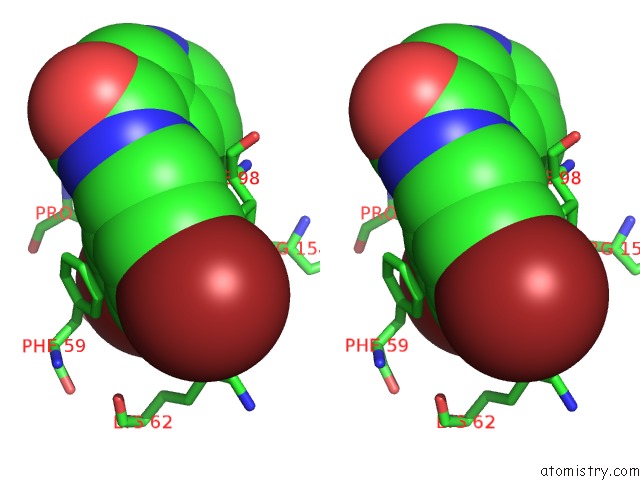
Bromine binding site 4 out of 4 in 2glp
Go back to
Bromine binding site 4 out
of 4 in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) From Helicobacter Pylori Complexed with Compound 1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 4 of Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) From Helicobacter Pylori Complexed with Compound 1 within 5.0Å range:
|
Reference:
L.Zhang,
W.Liu,
T.Hu,
L.Du,
C.Luo,
K.Chen,
X.Shen,
H.Jiang.
Structural Basis For Catalytic and Inhibitory Mechanisms of Beta-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz). J.Biol.Chem. V. 283 5370 2008.
ISSN: ISSN 0021-9258
PubMed: 18093984
DOI: 10.1074/JBC.M705566200
Page generated: Mon Jul 7 04:17:35 2025
ISSN: ISSN 0021-9258
PubMed: 18093984
DOI: 10.1074/JBC.M705566200
Last articles
F in 4G1WF in 4G16
F in 4FXY
F in 4FXQ
F in 4FZF
F in 4FP1
F in 4FVX
F in 4FV1
F in 4FV9
F in 4FV3