Bromine »
PDB 2hc0-2jkl »
2htl »
Bromine in PDB 2htl: Structure of the Escherichia Coli Clc Chloride Channel Y445F Mutant and Fab Complex
Protein crystallography data
The structure of Structure of the Escherichia Coli Clc Chloride Channel Y445F Mutant and Fab Complex, PDB code: 2htl
was solved by
A.Accardi,
S.Lobet,
C.Williams,
C.Miller,
R.Dutzler,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.00 / 3.40 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 232.525, 98.860, 171.804, 90.00, 131.54, 90.00 |
| R / Rfree (%) | 25.9 / 30.2 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Structure of the Escherichia Coli Clc Chloride Channel Y445F Mutant and Fab Complex
(pdb code 2htl). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 4 binding sites of Bromine where determined in the Structure of the Escherichia Coli Clc Chloride Channel Y445F Mutant and Fab Complex, PDB code: 2htl:
Jump to Bromine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Bromine where determined in the Structure of the Escherichia Coli Clc Chloride Channel Y445F Mutant and Fab Complex, PDB code: 2htl:
Jump to Bromine binding site number: 1; 2; 3; 4;
Bromine binding site 1 out of 4 in 2htl
Go back to
Bromine binding site 1 out
of 4 in the Structure of the Escherichia Coli Clc Chloride Channel Y445F Mutant and Fab Complex
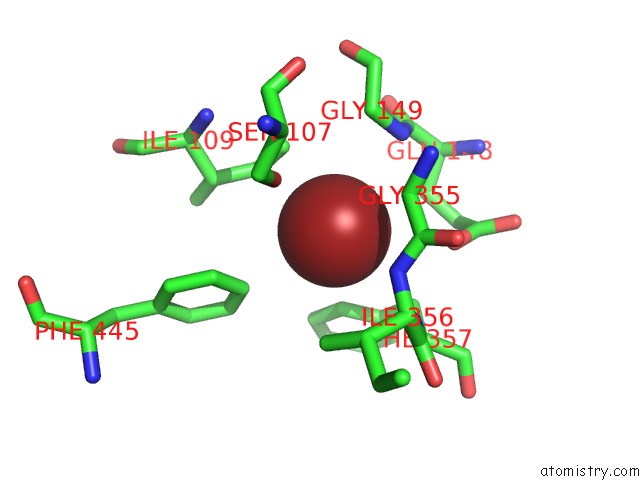
Mono view
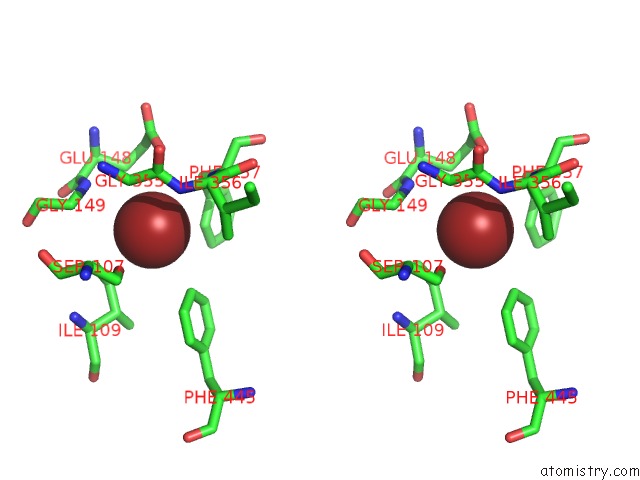
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Structure of the Escherichia Coli Clc Chloride Channel Y445F Mutant and Fab Complex within 5.0Å range:
|
Bromine binding site 2 out of 4 in 2htl
Go back to
Bromine binding site 2 out
of 4 in the Structure of the Escherichia Coli Clc Chloride Channel Y445F Mutant and Fab Complex
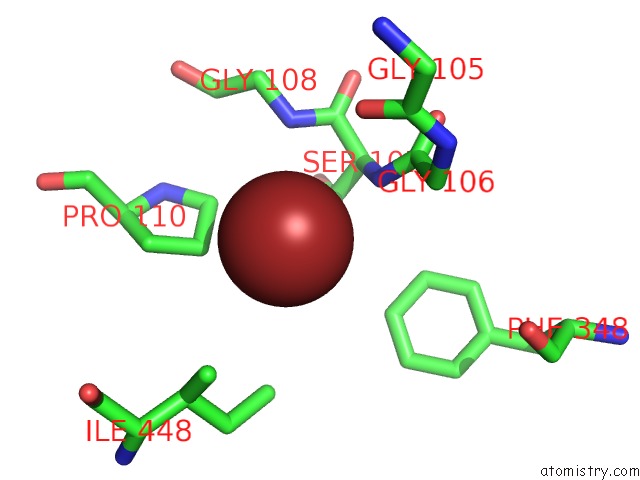
Mono view
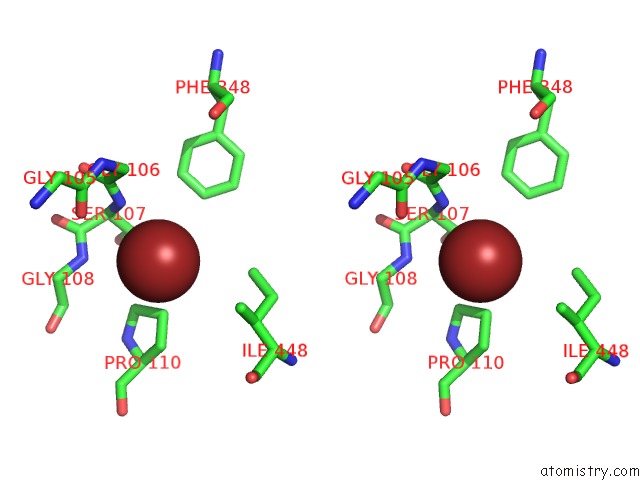
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Structure of the Escherichia Coli Clc Chloride Channel Y445F Mutant and Fab Complex within 5.0Å range:
|
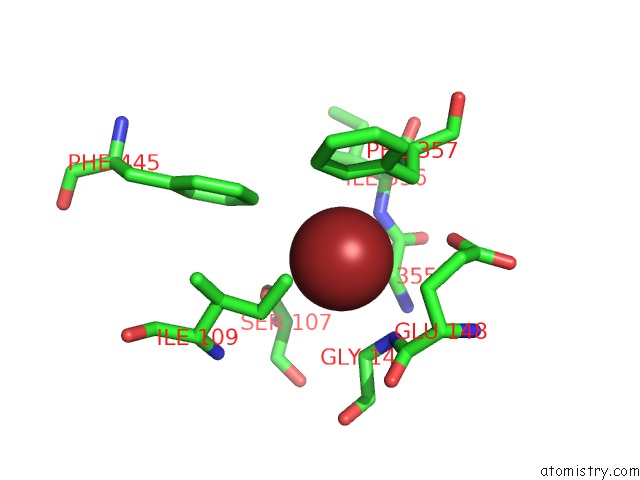
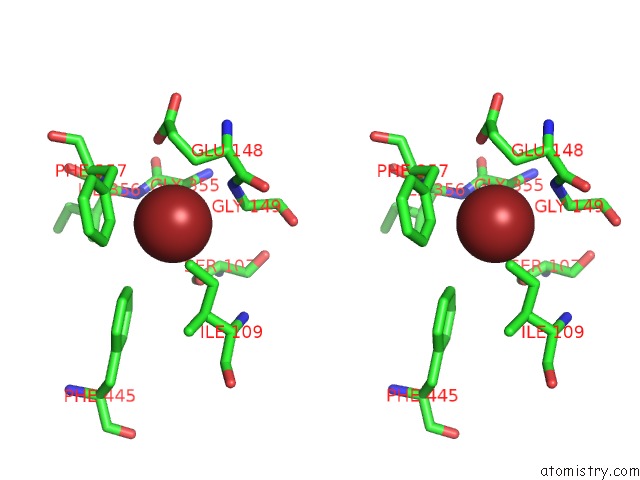
Bromine binding site 3 out of 4 in 2htl
Go back to
Bromine binding site 3 out
of 4 in the Structure of the Escherichia Coli Clc Chloride Channel Y445F Mutant and Fab Complex
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of Structure of the Escherichia Coli Clc Chloride Channel Y445F Mutant and Fab Complex within 5.0Å range:
|
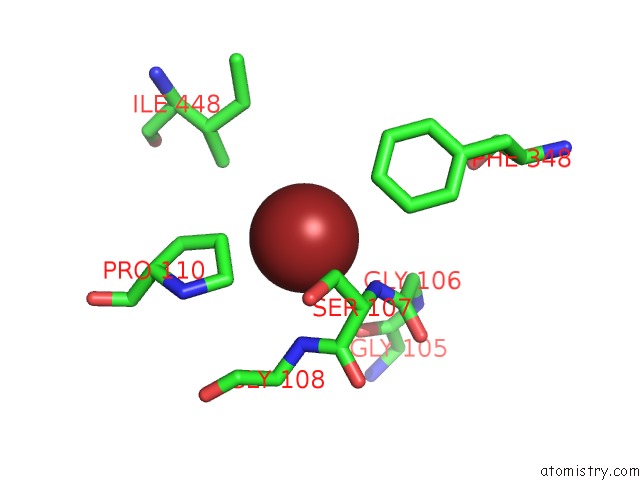
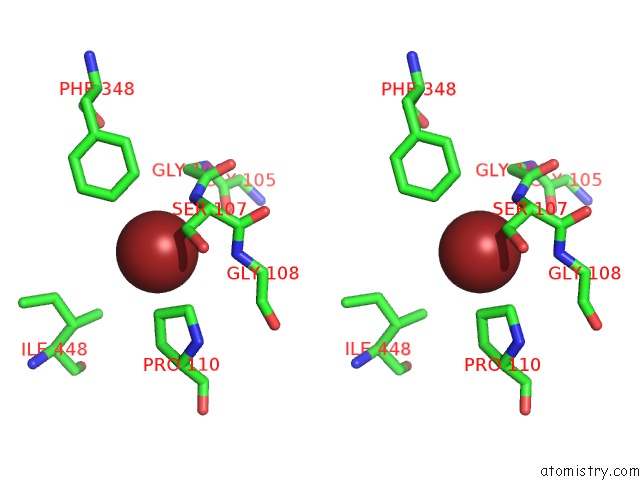
Bromine binding site 4 out of 4 in 2htl
Go back to
Bromine binding site 4 out
of 4 in the Structure of the Escherichia Coli Clc Chloride Channel Y445F Mutant and Fab Complex
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 4 of Structure of the Escherichia Coli Clc Chloride Channel Y445F Mutant and Fab Complex within 5.0Å range:
|
Reference:
A.Accardi,
S.Lobet,
C.Williams,
C.Miller,
R.Dutzler.
Synergism Between Halide Binding and Proton Transport in A Clc-Type Exchanger. J.Mol.Biol. V. 362 691 2006.
ISSN: ISSN 0022-2836
PubMed: 16949616
DOI: 10.1016/J.JMB.2006.07.081
Page generated: Mon Jul 7 04:21:21 2025
ISSN: ISSN 0022-2836
PubMed: 16949616
DOI: 10.1016/J.JMB.2006.07.081
Last articles
Br in 6E41Br in 6E40
Br in 6DV1
Br in 6E3T
Br in 6DRQ
Br in 6DLX
Br in 6DDA
Br in 6D6P
Br in 6DJH
Br in 6DFO