Bromine »
PDB 3jum-3m96 »
3khj »
Bromine in PDB 3khj: C. Parvum Inosine Monophosphate Dehydrogenase Bound By Inhibitor C64
Enzymatic activity of C. Parvum Inosine Monophosphate Dehydrogenase Bound By Inhibitor C64
All present enzymatic activity of C. Parvum Inosine Monophosphate Dehydrogenase Bound By Inhibitor C64:
1.1.1.205;
1.1.1.205;
Protein crystallography data
The structure of C. Parvum Inosine Monophosphate Dehydrogenase Bound By Inhibitor C64, PDB code: 3khj
was solved by
I.S.Macpherson,
L.K.Hedstrom,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.14 / 2.80 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 83.481, 166.141, 101.289, 90.00, 105.14, 90.00 |
| R / Rfree (%) | 22.4 / 26.6 |
Bromine Binding Sites:
The binding sites of Bromine atom in the C. Parvum Inosine Monophosphate Dehydrogenase Bound By Inhibitor C64
(pdb code 3khj). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 3 binding sites of Bromine where determined in the C. Parvum Inosine Monophosphate Dehydrogenase Bound By Inhibitor C64, PDB code: 3khj:
Jump to Bromine binding site number: 1; 2; 3;
In total 3 binding sites of Bromine where determined in the C. Parvum Inosine Monophosphate Dehydrogenase Bound By Inhibitor C64, PDB code: 3khj:
Jump to Bromine binding site number: 1; 2; 3;
Bromine binding site 1 out of 3 in 3khj
Go back to
Bromine binding site 1 out
of 3 in the C. Parvum Inosine Monophosphate Dehydrogenase Bound By Inhibitor C64
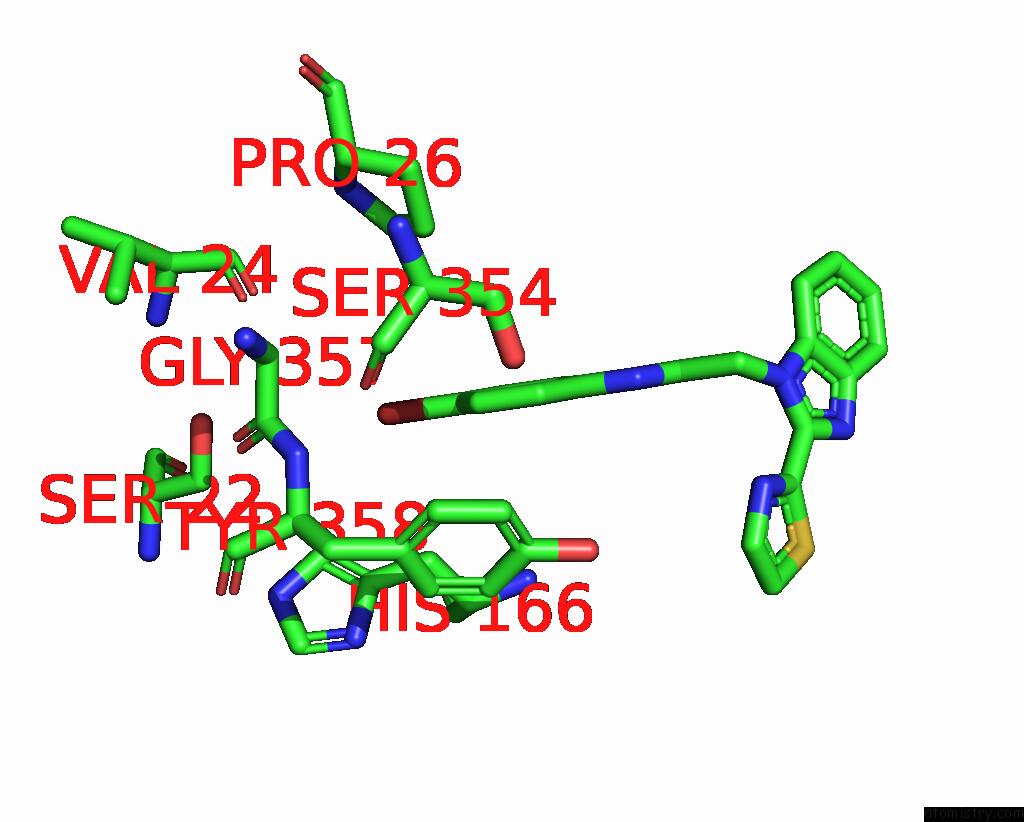
Mono view
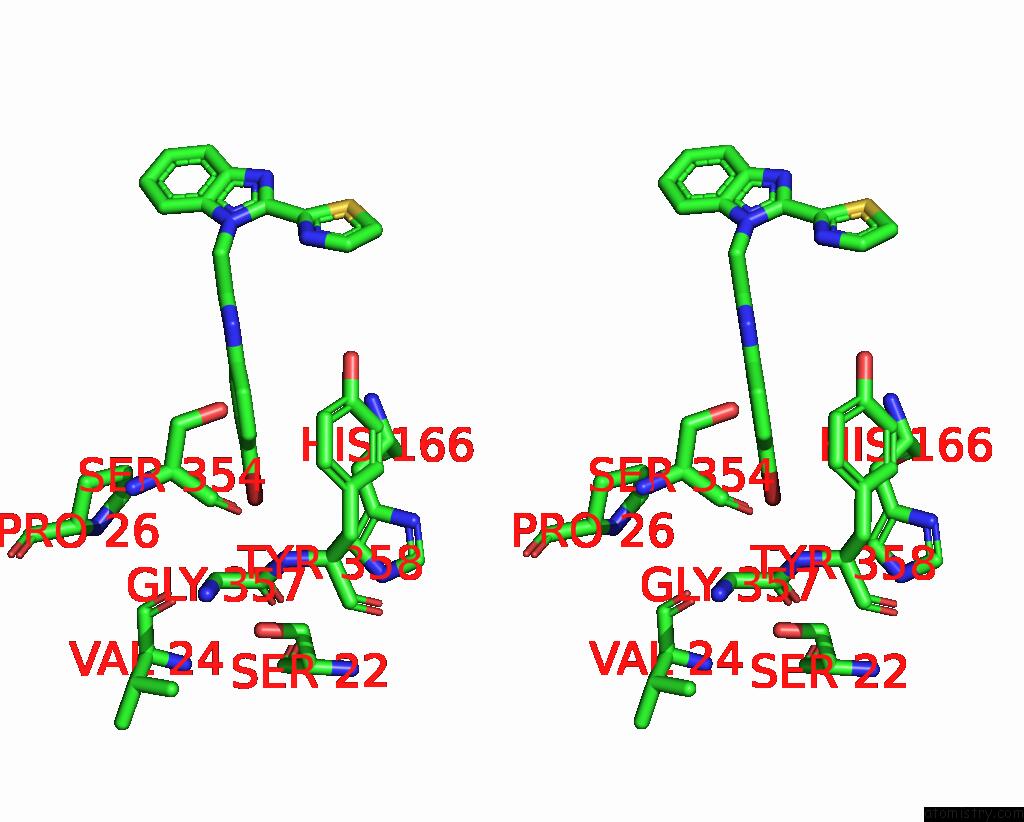
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of C. Parvum Inosine Monophosphate Dehydrogenase Bound By Inhibitor C64 within 5.0Å range:
|
Bromine binding site 2 out of 3 in 3khj
Go back to
Bromine binding site 2 out
of 3 in the C. Parvum Inosine Monophosphate Dehydrogenase Bound By Inhibitor C64
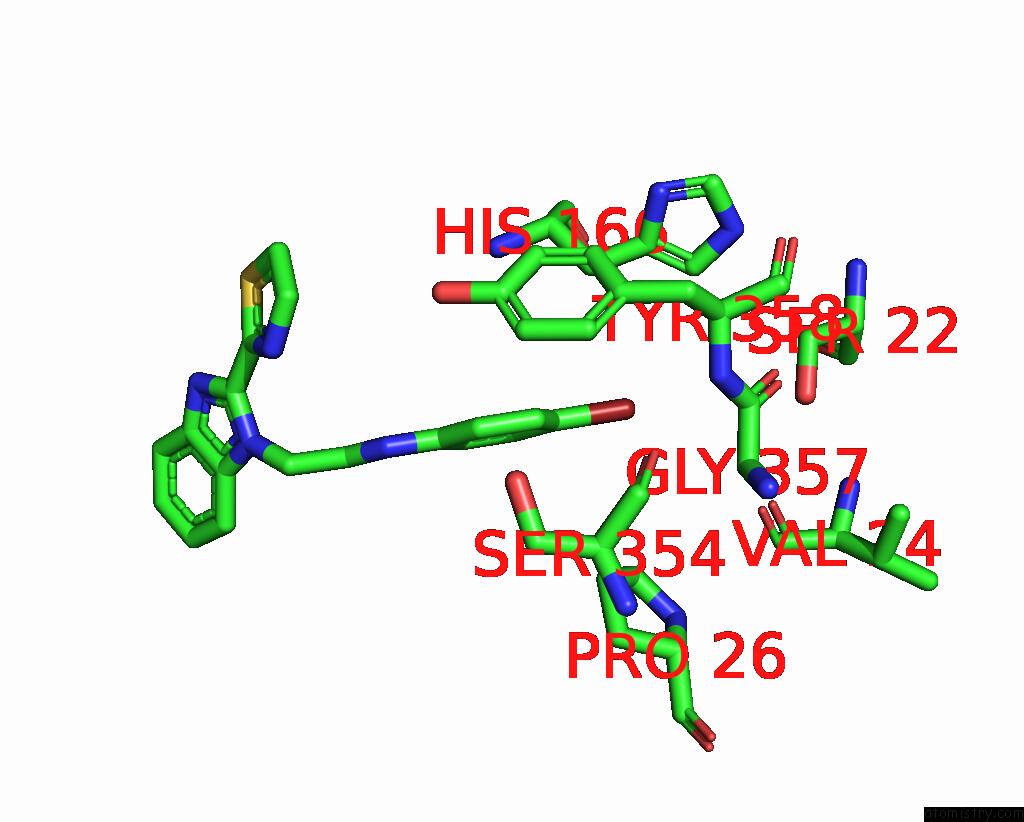
Mono view
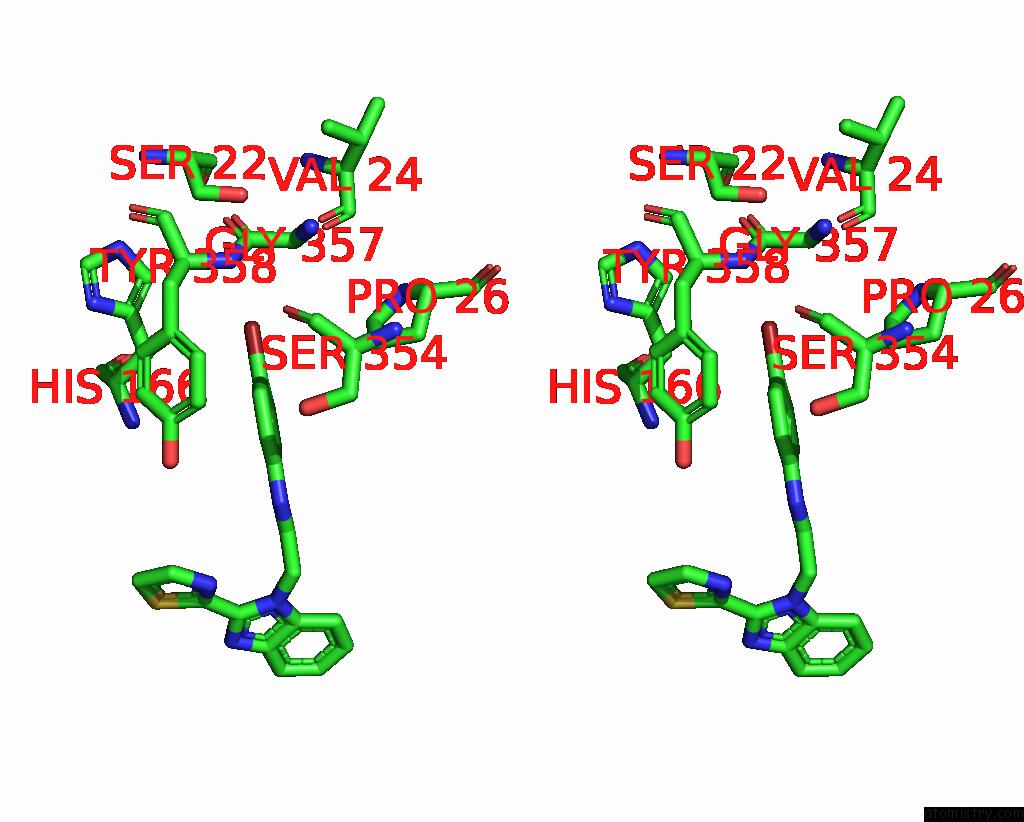
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of C. Parvum Inosine Monophosphate Dehydrogenase Bound By Inhibitor C64 within 5.0Å range:
|
Bromine binding site 3 out of 3 in 3khj
Go back to
Bromine binding site 3 out
of 3 in the C. Parvum Inosine Monophosphate Dehydrogenase Bound By Inhibitor C64
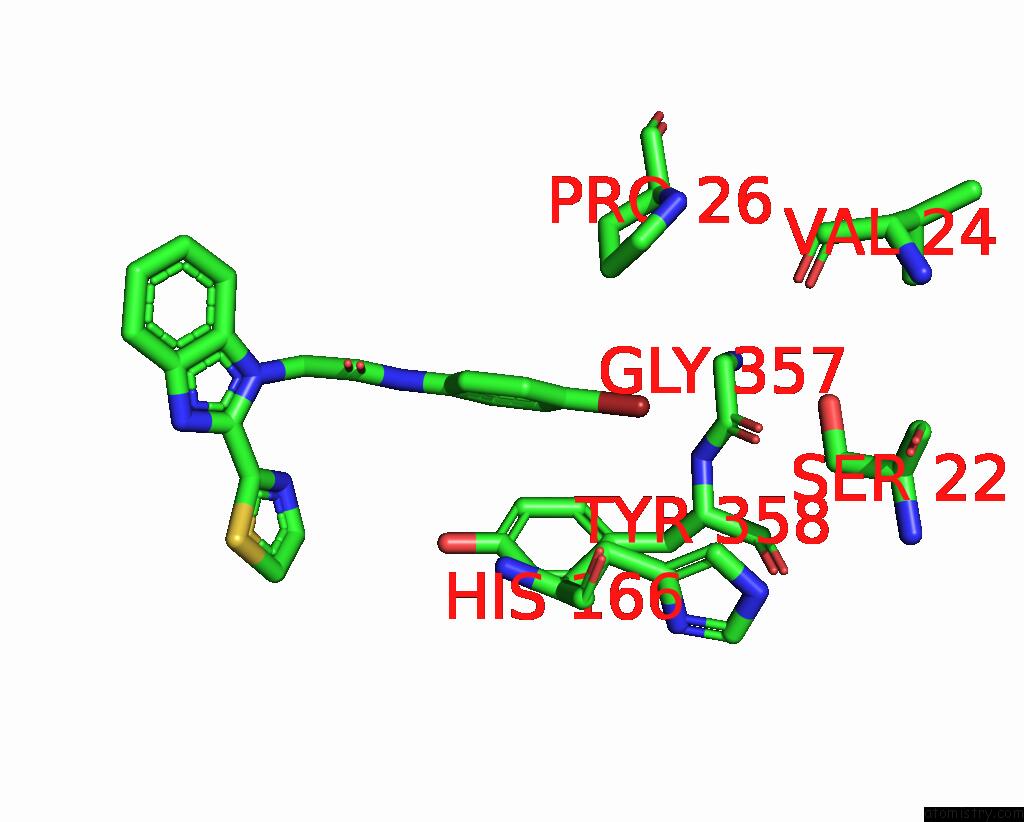
Mono view
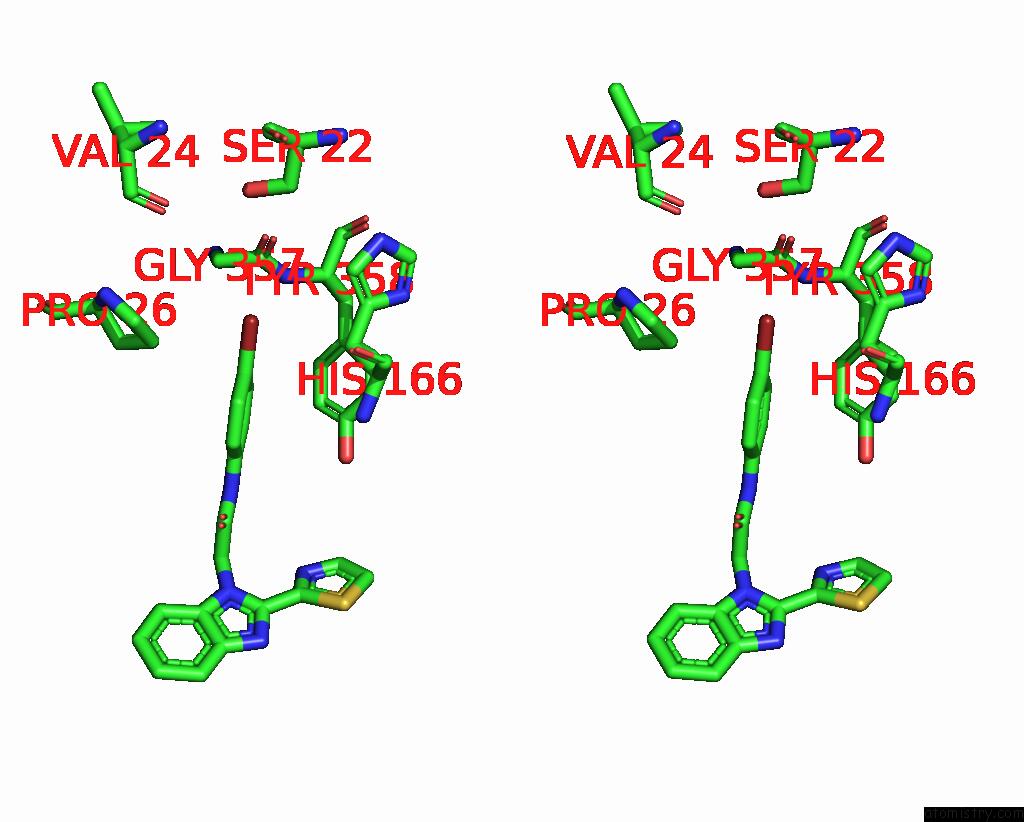
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of C. Parvum Inosine Monophosphate Dehydrogenase Bound By Inhibitor C64 within 5.0Å range:
|
Reference:
I.S.Macpherson,
S.Kirubakaran,
S.K.Gorla,
T.V.Riera,
J.A.D'aquino,
M.Zhang,
G.D.Cuny,
L.Hedstrom.
The Structural Basis of Cryptosporidium -Specific Imp Dehydrogenase Inhibitor Selectivity. J.Am.Chem.Soc. V. 132 1230 2010.
ISSN: ISSN 0002-7863
PubMed: 20052976
DOI: 10.1021/JA909947A
Page generated: Wed Jul 10 19:51:03 2024
ISSN: ISSN 0002-7863
PubMed: 20052976
DOI: 10.1021/JA909947A
Last articles
Ba in 3GOMBa in 3GOJ
Ba in 2YN6
Ba in 3FQB
Ba in 3E8F
Ba in 2YDH
Ba in 3E8V
Ba in 3ATA
Ba in 3AT8
Ba in 3AA7