Bromine »
PDB 3ma6-3oji »
3ns2 »
Bromine in PDB 3ns2: High-Resolution Structure of Pyrabactin-Bound PYL2
Protein crystallography data
The structure of High-Resolution Structure of Pyrabactin-Bound PYL2, PDB code: 3ns2
was solved by
Q.Hao,
P.Yin,
C.Yan,
X.Yuan,
J.Wang,
N.Yan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.38 / 1.63 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 62.520, 104.600, 184.300, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.6 / 21.8 |
Bromine Binding Sites:
The binding sites of Bromine atom in the High-Resolution Structure of Pyrabactin-Bound PYL2
(pdb code 3ns2). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 3 binding sites of Bromine where determined in the High-Resolution Structure of Pyrabactin-Bound PYL2, PDB code: 3ns2:
Jump to Bromine binding site number: 1; 2; 3;
In total 3 binding sites of Bromine where determined in the High-Resolution Structure of Pyrabactin-Bound PYL2, PDB code: 3ns2:
Jump to Bromine binding site number: 1; 2; 3;
Bromine binding site 1 out of 3 in 3ns2
Go back to
Bromine binding site 1 out
of 3 in the High-Resolution Structure of Pyrabactin-Bound PYL2
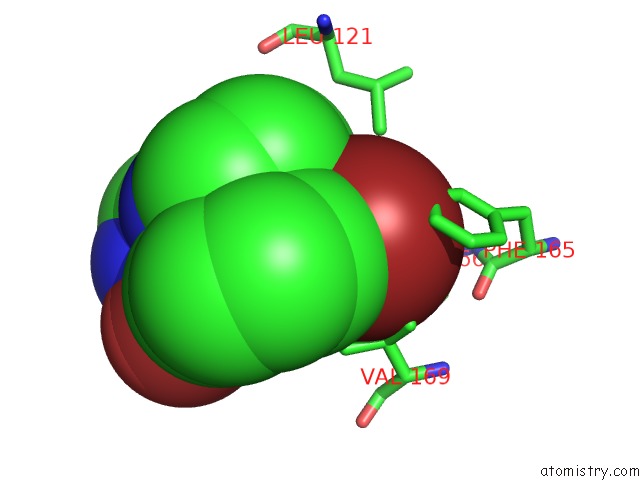
Mono view
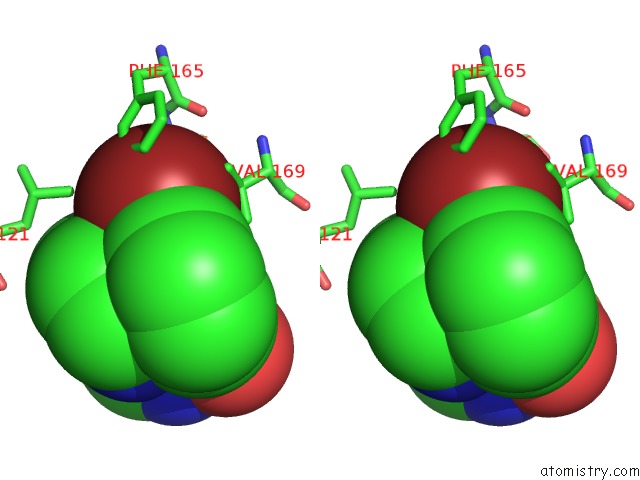
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of High-Resolution Structure of Pyrabactin-Bound PYL2 within 5.0Å range:
|
Bromine binding site 2 out of 3 in 3ns2
Go back to
Bromine binding site 2 out
of 3 in the High-Resolution Structure of Pyrabactin-Bound PYL2
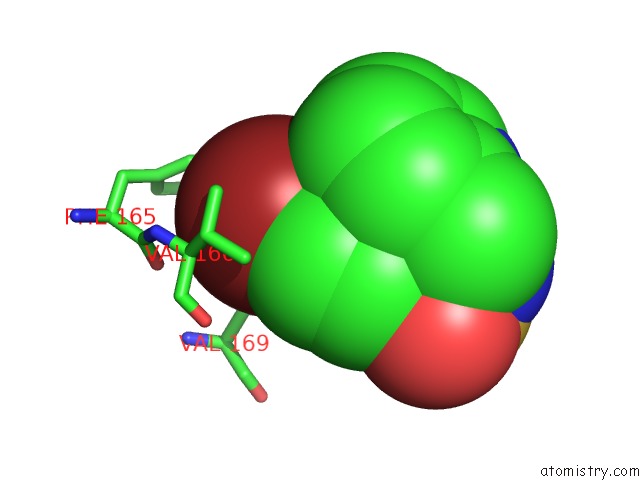
Mono view
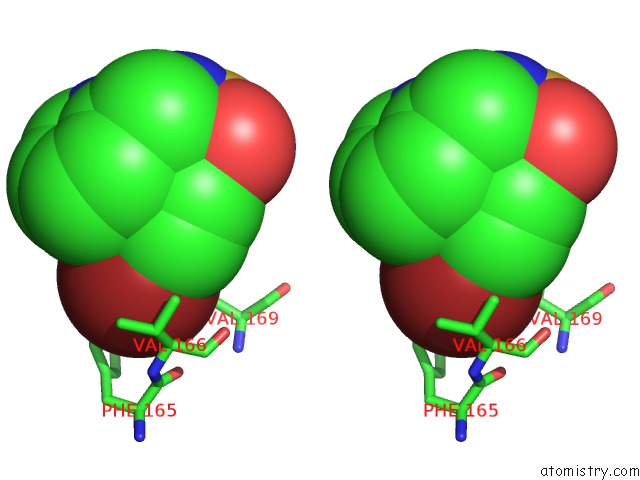
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of High-Resolution Structure of Pyrabactin-Bound PYL2 within 5.0Å range:
|
Bromine binding site 3 out of 3 in 3ns2
Go back to
Bromine binding site 3 out
of 3 in the High-Resolution Structure of Pyrabactin-Bound PYL2
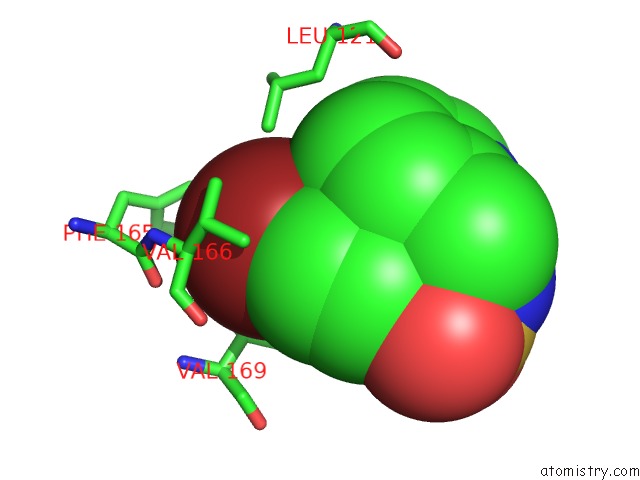
Mono view
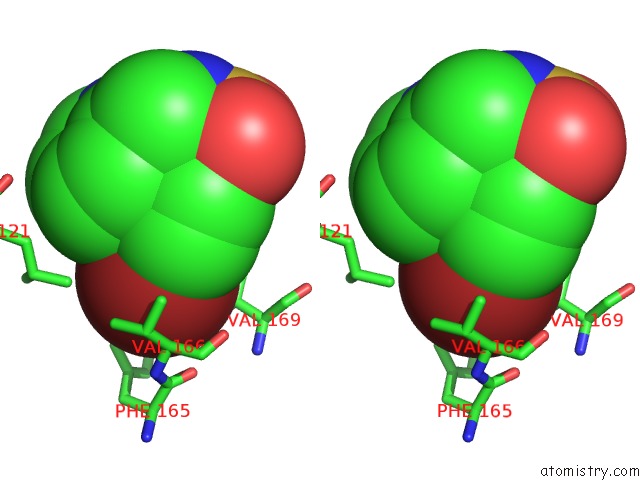
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of High-Resolution Structure of Pyrabactin-Bound PYL2 within 5.0Å range:
|
Reference:
X.Yuan,
P.Yin,
Q.Hao,
C.Yan,
J.Wang,
N.Yan.
Single Amino Acid Alteration Between Valine and Isoleucine Determines the Distinct Pyrabactin Selectivity By PYL1 and PYL2 J.Biol.Chem. V. 285 28953 2010.
ISSN: ISSN 0021-9258
PubMed: 20630864
DOI: 10.1074/JBC.M110.160192
Page generated: Mon Jul 7 05:43:28 2025
ISSN: ISSN 0021-9258
PubMed: 20630864
DOI: 10.1074/JBC.M110.160192
Last articles
F in 7JTOF in 7JOM
F in 7JT4
F in 7JRA
F in 7JR6
F in 7JPW
F in 7JPL
F in 7JPK
F in 7JL3
F in 7JN3