Bromine »
PDB 3ok5-3r1e »
3pba »
Bromine in PDB 3pba: Crystal Structure of Ppargamma Ligand Binding Domain in Complex with Monosulfate Tetrabromo-Bisphenol A (Monotbbpa)
Protein crystallography data
The structure of Crystal Structure of Ppargamma Ligand Binding Domain in Complex with Monosulfate Tetrabromo-Bisphenol A (Monotbbpa), PDB code: 3pba
was solved by
A.Le Maire,
W.Bourguet,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.93 / 2.30 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 93.371, 61.564, 119.074, 90.00, 102.89, 90.00 |
| R / Rfree (%) | 18.8 / 25.5 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Crystal Structure of Ppargamma Ligand Binding Domain in Complex with Monosulfate Tetrabromo-Bisphenol A (Monotbbpa)
(pdb code 3pba). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 4 binding sites of Bromine where determined in the Crystal Structure of Ppargamma Ligand Binding Domain in Complex with Monosulfate Tetrabromo-Bisphenol A (Monotbbpa), PDB code: 3pba:
Jump to Bromine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Bromine where determined in the Crystal Structure of Ppargamma Ligand Binding Domain in Complex with Monosulfate Tetrabromo-Bisphenol A (Monotbbpa), PDB code: 3pba:
Jump to Bromine binding site number: 1; 2; 3; 4;
Bromine binding site 1 out of 4 in 3pba
Go back to
Bromine binding site 1 out
of 4 in the Crystal Structure of Ppargamma Ligand Binding Domain in Complex with Monosulfate Tetrabromo-Bisphenol A (Monotbbpa)
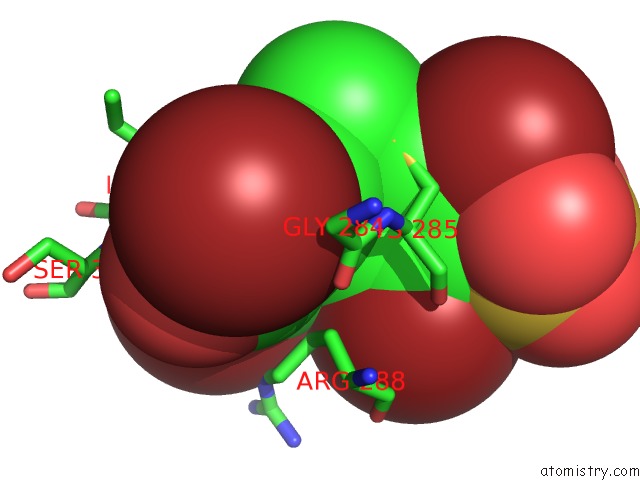
Mono view
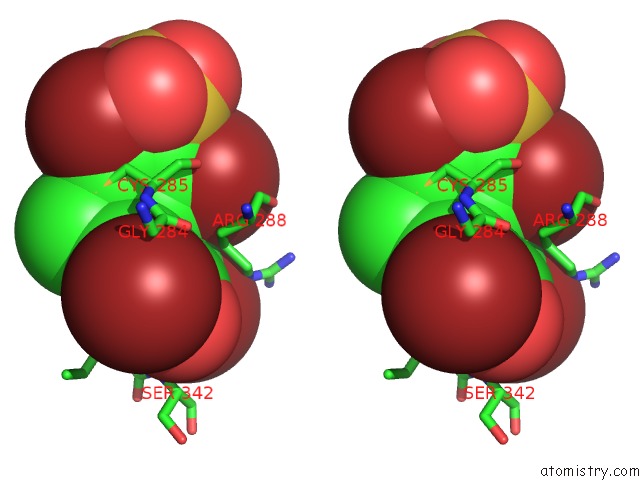
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Crystal Structure of Ppargamma Ligand Binding Domain in Complex with Monosulfate Tetrabromo-Bisphenol A (Monotbbpa) within 5.0Å range:
|
Bromine binding site 2 out of 4 in 3pba
Go back to
Bromine binding site 2 out
of 4 in the Crystal Structure of Ppargamma Ligand Binding Domain in Complex with Monosulfate Tetrabromo-Bisphenol A (Monotbbpa)
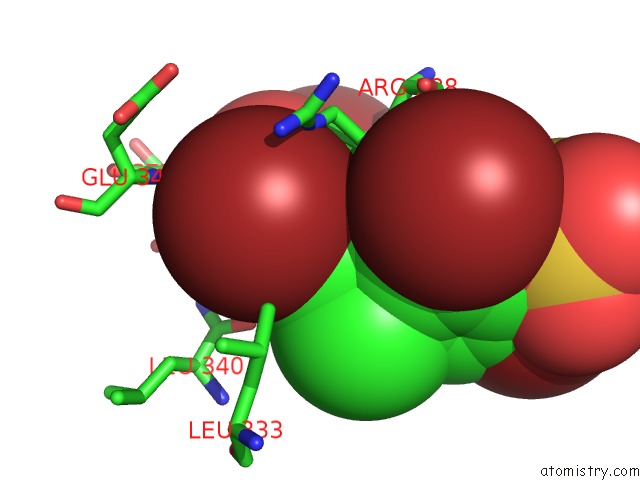
Mono view
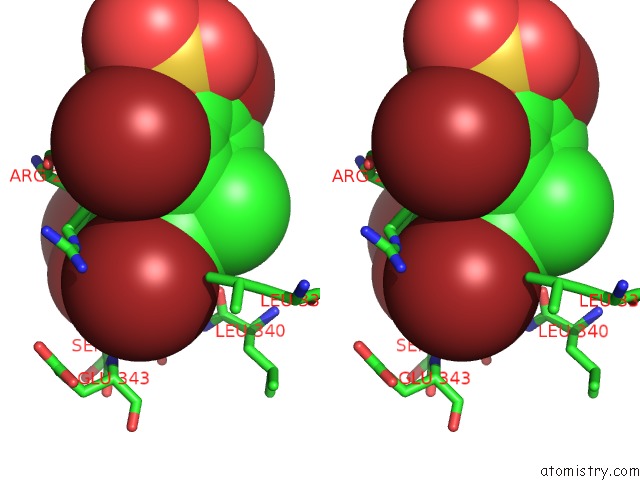
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Crystal Structure of Ppargamma Ligand Binding Domain in Complex with Monosulfate Tetrabromo-Bisphenol A (Monotbbpa) within 5.0Å range:
|
Bromine binding site 3 out of 4 in 3pba
Go back to
Bromine binding site 3 out
of 4 in the Crystal Structure of Ppargamma Ligand Binding Domain in Complex with Monosulfate Tetrabromo-Bisphenol A (Monotbbpa)
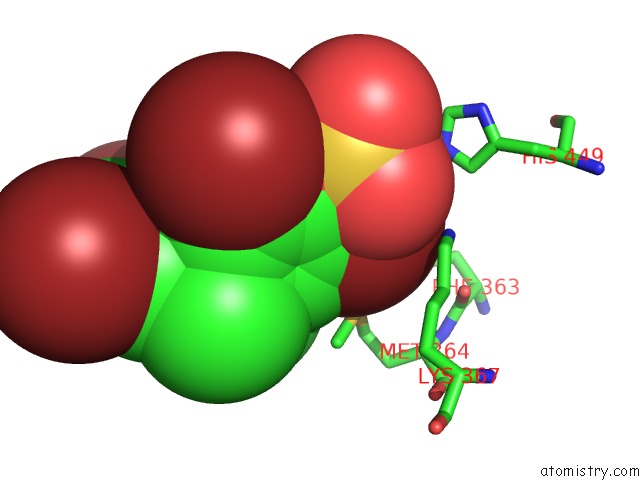
Mono view
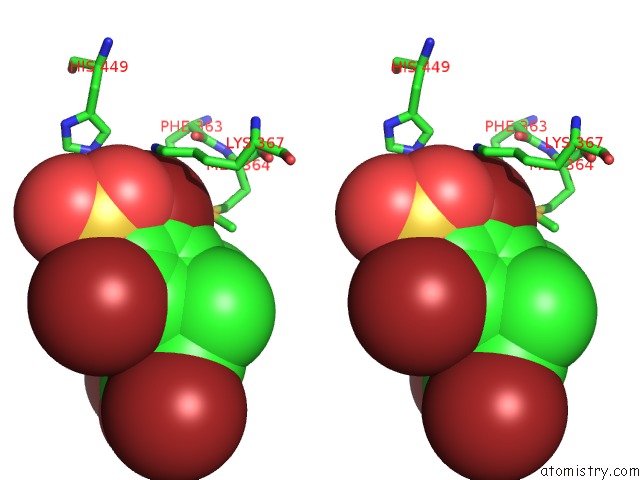
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of Crystal Structure of Ppargamma Ligand Binding Domain in Complex with Monosulfate Tetrabromo-Bisphenol A (Monotbbpa) within 5.0Å range:
|
Bromine binding site 4 out of 4 in 3pba
Go back to
Bromine binding site 4 out
of 4 in the Crystal Structure of Ppargamma Ligand Binding Domain in Complex with Monosulfate Tetrabromo-Bisphenol A (Monotbbpa)
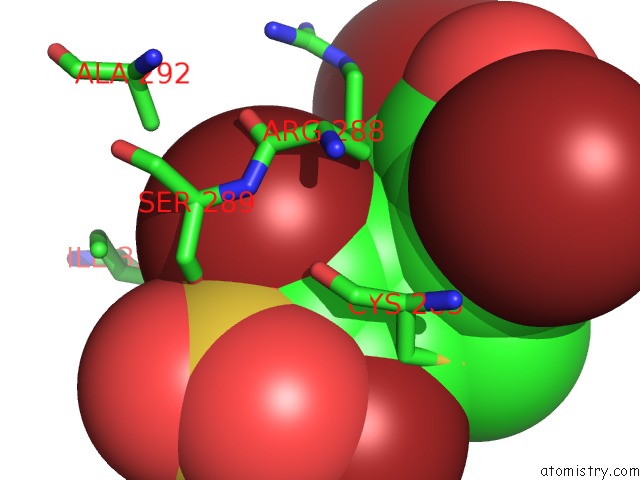
Mono view
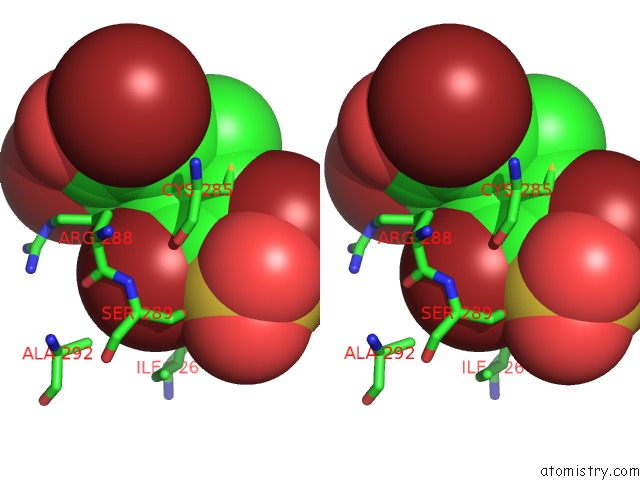
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 4 of Crystal Structure of Ppargamma Ligand Binding Domain in Complex with Monosulfate Tetrabromo-Bisphenol A (Monotbbpa) within 5.0Å range:
|
Reference:
A.Riu,
A.Le Maire,
M.Grimaldi,
M.Audebert,
A.Hillenweck,
W.Bourguet,
P.Balaguer,
D.Zalko.
Characterization of Novel Ligands of Er{Alpha}, Er{Beta}, and Ppar{Gamma}: the Case of Halogenated Bisphenol A and Their Conjugated Metabolites. Toxicol Sci V. 122 372 2011.
ISSN: ISSN 1096-0929
PubMed: 21622942
DOI: 10.1093/TOXSCI/KFR132
Page generated: Mon Jul 7 05:48:02 2025
ISSN: ISSN 1096-0929
PubMed: 21622942
DOI: 10.1093/TOXSCI/KFR132
Last articles
Br in 8KI3Br in 8K4H
Br in 8KHF
Br in 8KH6
Br in 8KE4
Br in 8KE1
Br in 8K2M
Br in 8K9L
Br in 8K6D
Br in 8JV5