Bromine »
PDB 4d4j-4g6v »
4dnf »
Bromine in PDB 4dnf: Crystal Structure of the Cftr Inhibitory Factor Cif with the E153Q Mutation Adducted with the Epibromohydrin Hydrolysis Intermediate
Protein crystallography data
The structure of Crystal Structure of the Cftr Inhibitory Factor Cif with the E153Q Mutation Adducted with the Epibromohydrin Hydrolysis Intermediate, PDB code: 4dnf
was solved by
C.D.Bahl,
D.R.Madden,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 38.54 / 1.80 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 168.147, 83.959, 89.536, 90.00, 100.31, 90.00 |
| R / Rfree (%) | 18.9 / 22.4 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Crystal Structure of the Cftr Inhibitory Factor Cif with the E153Q Mutation Adducted with the Epibromohydrin Hydrolysis Intermediate
(pdb code 4dnf). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 4 binding sites of Bromine where determined in the Crystal Structure of the Cftr Inhibitory Factor Cif with the E153Q Mutation Adducted with the Epibromohydrin Hydrolysis Intermediate, PDB code: 4dnf:
Jump to Bromine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Bromine where determined in the Crystal Structure of the Cftr Inhibitory Factor Cif with the E153Q Mutation Adducted with the Epibromohydrin Hydrolysis Intermediate, PDB code: 4dnf:
Jump to Bromine binding site number: 1; 2; 3; 4;
Bromine binding site 1 out of 4 in 4dnf
Go back to
Bromine binding site 1 out
of 4 in the Crystal Structure of the Cftr Inhibitory Factor Cif with the E153Q Mutation Adducted with the Epibromohydrin Hydrolysis Intermediate
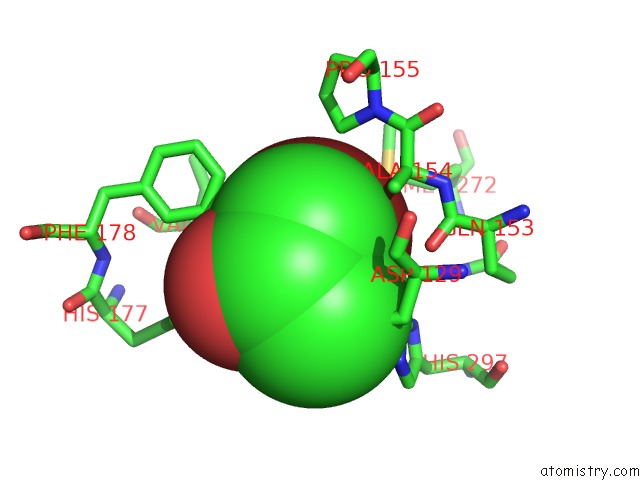
Mono view
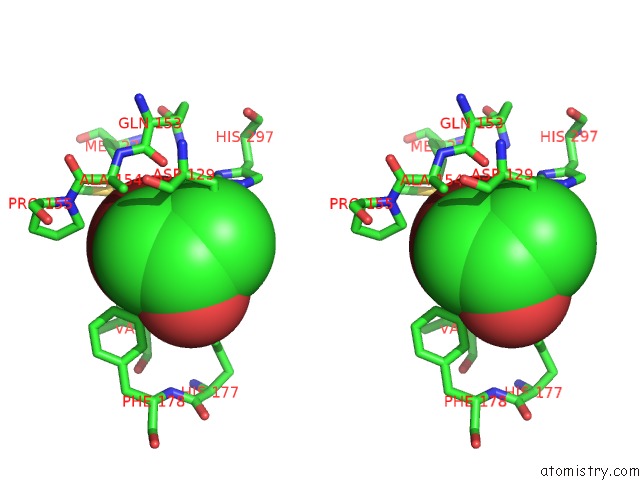
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Crystal Structure of the Cftr Inhibitory Factor Cif with the E153Q Mutation Adducted with the Epibromohydrin Hydrolysis Intermediate within 5.0Å range:
|
Bromine binding site 2 out of 4 in 4dnf
Go back to
Bromine binding site 2 out
of 4 in the Crystal Structure of the Cftr Inhibitory Factor Cif with the E153Q Mutation Adducted with the Epibromohydrin Hydrolysis Intermediate
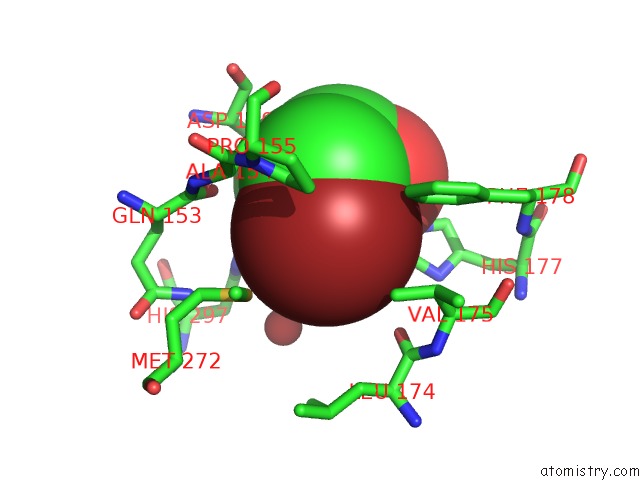
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Crystal Structure of the Cftr Inhibitory Factor Cif with the E153Q Mutation Adducted with the Epibromohydrin Hydrolysis Intermediate within 5.0Å range:
|
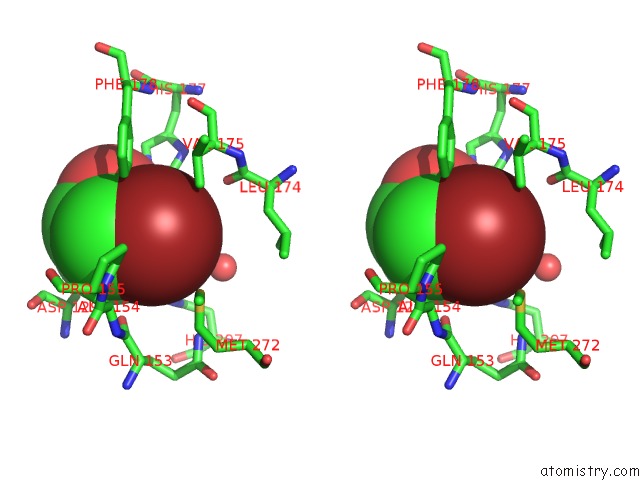
Bromine binding site 3 out of 4 in 4dnf
Go back to
Bromine binding site 3 out
of 4 in the Crystal Structure of the Cftr Inhibitory Factor Cif with the E153Q Mutation Adducted with the Epibromohydrin Hydrolysis Intermediate
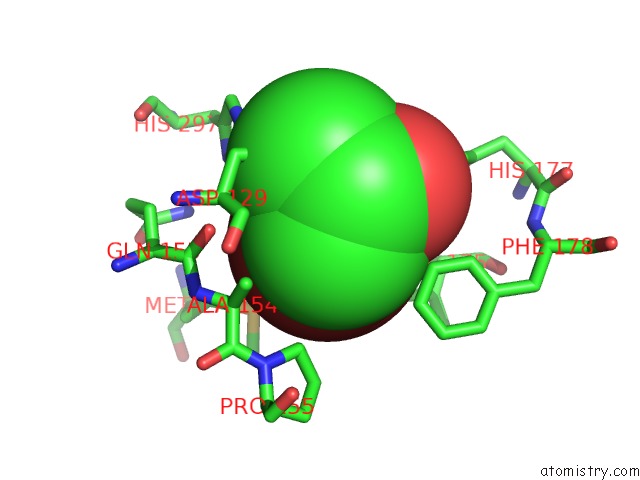
Mono view
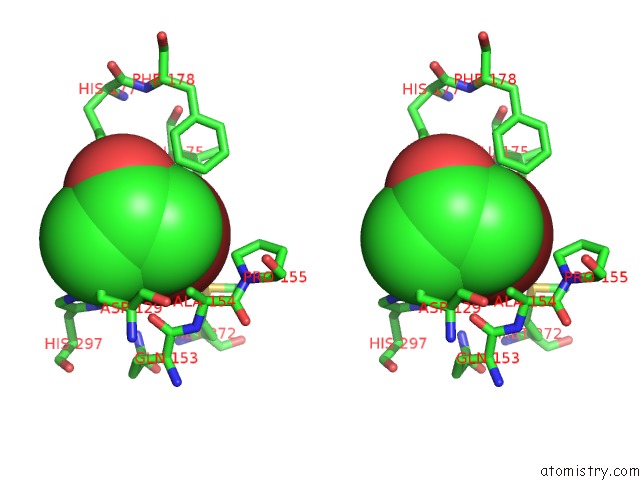
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of Crystal Structure of the Cftr Inhibitory Factor Cif with the E153Q Mutation Adducted with the Epibromohydrin Hydrolysis Intermediate within 5.0Å range:
|
Bromine binding site 4 out of 4 in 4dnf
Go back to
Bromine binding site 4 out
of 4 in the Crystal Structure of the Cftr Inhibitory Factor Cif with the E153Q Mutation Adducted with the Epibromohydrin Hydrolysis Intermediate
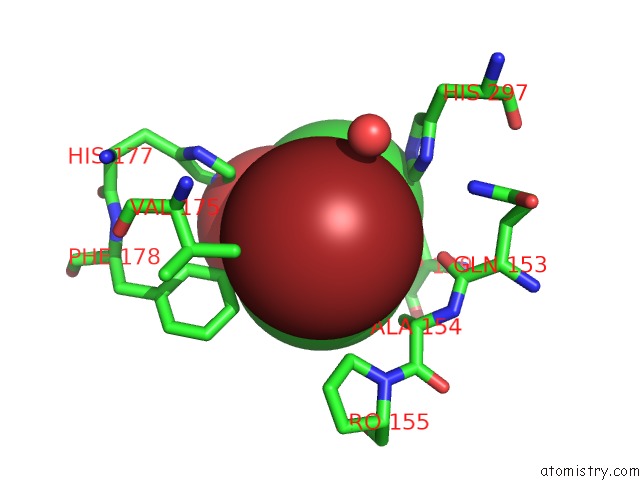
Mono view
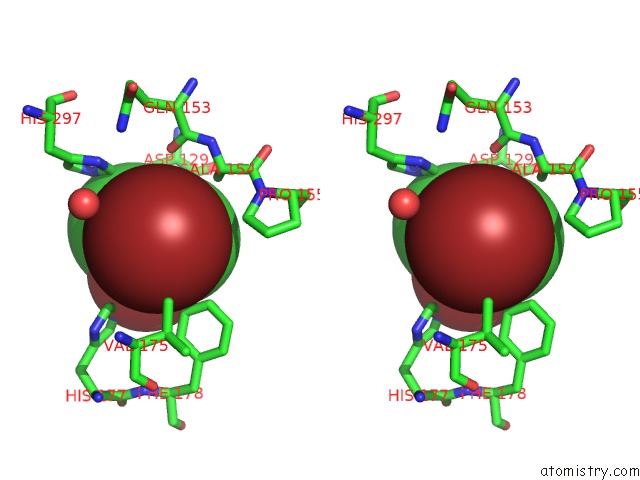
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 4 of Crystal Structure of the Cftr Inhibitory Factor Cif with the E153Q Mutation Adducted with the Epibromohydrin Hydrolysis Intermediate within 5.0Å range:
|
Reference:
C.D.Bahl,
K.L.Hvorecny,
C.Morisseau,
S.A.Gerber,
D.R.Madden.
Visualizing the Mechanism of Epoxide Hydrolysis By the Bacterial Virulence Enzyme Cif. Biochemistry V. 55 788 2016.
ISSN: ISSN 0006-2960
PubMed: 26752215
DOI: 10.1021/ACS.BIOCHEM.5B01229
Page generated: Mon Jul 7 06:27:25 2025
ISSN: ISSN 0006-2960
PubMed: 26752215
DOI: 10.1021/ACS.BIOCHEM.5B01229
Last articles
Br in 7ZH8Br in 8A9P
Br in 8A7B
Br in 7ZYO
Br in 7ZYR
Br in 8A7A
Br in 7ZYK
Br in 7ZY2
Br in 7ZYD
Br in 7ZSR