Bromine »
PDB 4kvh-4my6 »
4mp2 »
Bromine in PDB 4mp2: Crystal Structure of Pyruvate Dehydrogenase Kinase Isoform 2 in Complex with Inhibitor PA1
Enzymatic activity of Crystal Structure of Pyruvate Dehydrogenase Kinase Isoform 2 in Complex with Inhibitor PA1
All present enzymatic activity of Crystal Structure of Pyruvate Dehydrogenase Kinase Isoform 2 in Complex with Inhibitor PA1:
2.7.11.2;
2.7.11.2;
Protein crystallography data
The structure of Crystal Structure of Pyruvate Dehydrogenase Kinase Isoform 2 in Complex with Inhibitor PA1, PDB code: 4mp2
was solved by
W.J.Gui,
S.C.Tso,
J.L.Chuang,
C.Y.Wu,
X.Qi,
U.K.Tambar,
R.M.Wynn,
D.T.Chuang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 36.93 / 1.75 |
| Space group | I 41 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 110.323, 110.323, 229.523, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.4 / 21.9 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Crystal Structure of Pyruvate Dehydrogenase Kinase Isoform 2 in Complex with Inhibitor PA1
(pdb code 4mp2). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total only one binding site of Bromine was determined in the Crystal Structure of Pyruvate Dehydrogenase Kinase Isoform 2 in Complex with Inhibitor PA1, PDB code: 4mp2:
In total only one binding site of Bromine was determined in the Crystal Structure of Pyruvate Dehydrogenase Kinase Isoform 2 in Complex with Inhibitor PA1, PDB code: 4mp2:
Bromine binding site 1 out of 1 in 4mp2
Go back to
Bromine binding site 1 out
of 1 in the Crystal Structure of Pyruvate Dehydrogenase Kinase Isoform 2 in Complex with Inhibitor PA1
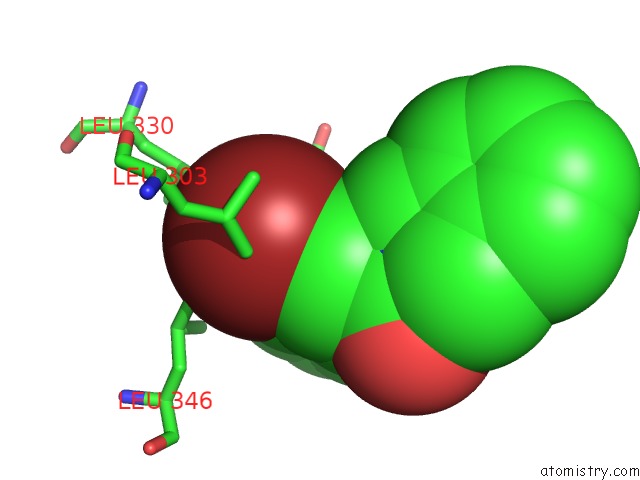
Mono view
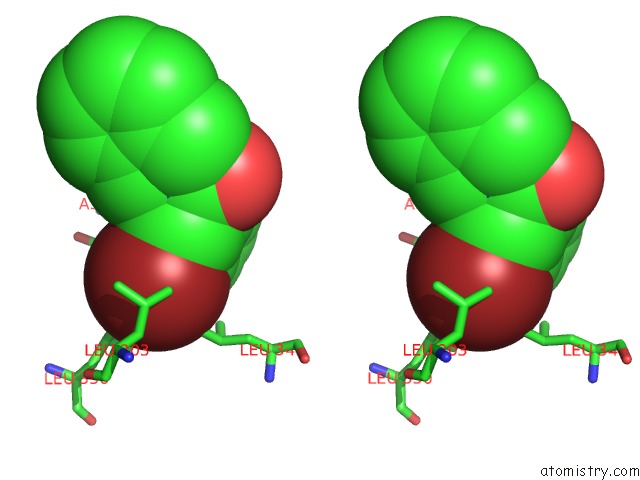
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Crystal Structure of Pyruvate Dehydrogenase Kinase Isoform 2 in Complex with Inhibitor PA1 within 5.0Å range:
|
Reference:
S.C.Tso,
X.Qi,
W.J.Gui,
C.Y.Wu,
J.L.Chuang,
I.Wernstedt-Asterholm,
L.K.Morlock,
K.R.Owens,
P.E.Scherer,
N.S.Williams,
U.K.Tambar,
R.M.Wynn,
D.T.Chuang.
Structure-Guided Development of Specific Pyruvate Dehydrogenase Kinase Inhibitors Targeting the Atp-Binding Pocket. J.Biol.Chem. V. 289 4432 2014.
ISSN: ISSN 0021-9258
PubMed: 24356970
DOI: 10.1074/JBC.M113.533885
Page generated: Mon Jul 7 07:06:25 2025
ISSN: ISSN 0021-9258
PubMed: 24356970
DOI: 10.1074/JBC.M113.533885
Last articles
Br in 8TWNBr in 8TSB
Br in 8TRN
Br in 8TSA
Br in 8TQD
Br in 8SA7
Br in 8TNV
Br in 8TG1
Br in 8SZ3
Br in 8TCV