Bromine »
PDB 4x6i-4z8b »
4xzi »
Bromine in PDB 4xzi: Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049
Enzymatic activity of Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049
All present enzymatic activity of Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049:
1.1.1.21;
1.1.1.21;
Protein crystallography data
The structure of Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049, PDB code: 4xzi
was solved by
A.Cousido-Siah,
F.X.Ruiz,
A.Mitschler,
M.Dominguez,
A.R.De Lera,
J.Farres,
X.Pares,
A.Podjarny,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.03 / 2.45 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.065, 84.780, 105.239, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.9 / 29 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049
(pdb code 4xzi). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 8 binding sites of Bromine where determined in the Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049, PDB code: 4xzi:
Jump to Bromine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Bromine where determined in the Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049, PDB code: 4xzi:
Jump to Bromine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Bromine binding site 1 out of 8 in 4xzi
Go back to
Bromine binding site 1 out
of 8 in the Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049
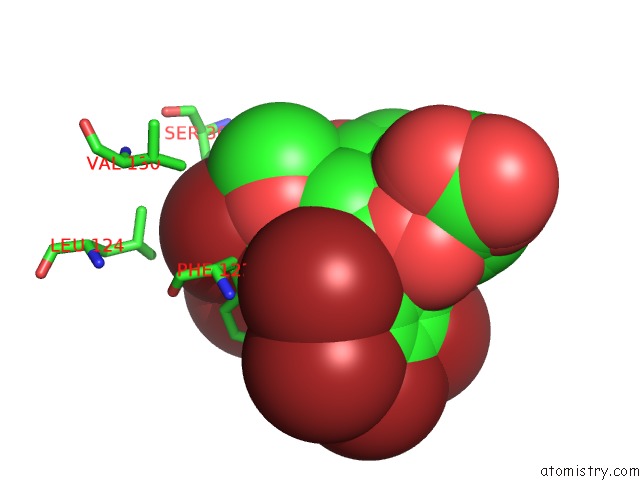
Mono view
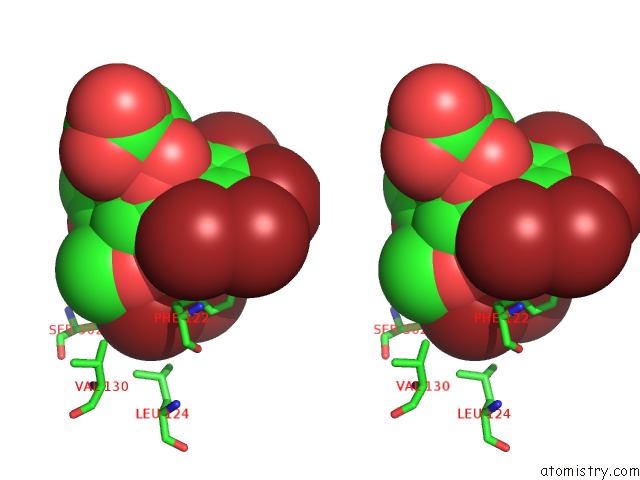
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049 within 5.0Å range:
|
Bromine binding site 2 out of 8 in 4xzi
Go back to
Bromine binding site 2 out
of 8 in the Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049
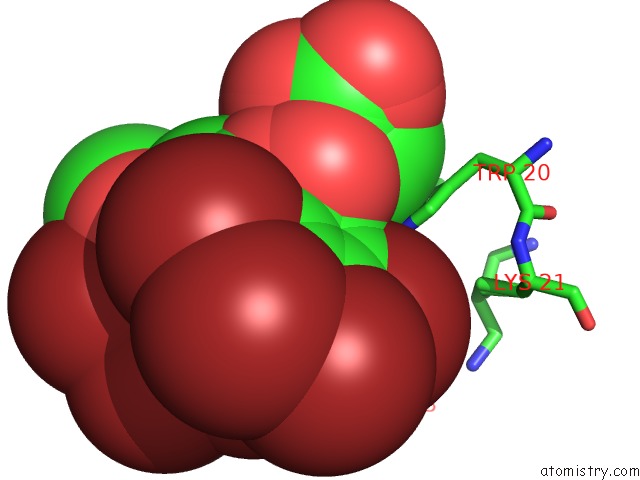
Mono view
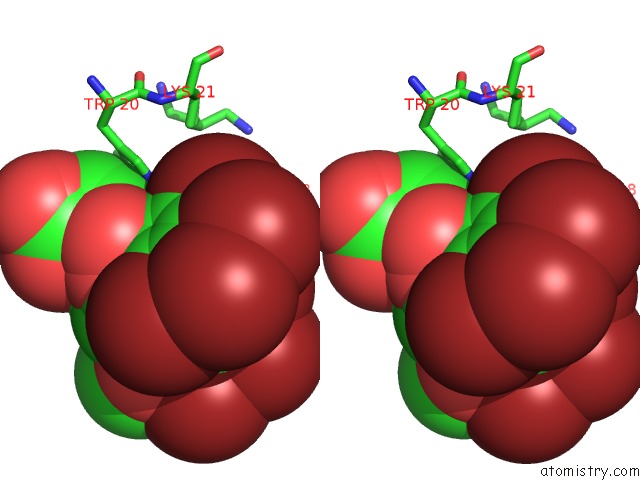
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049 within 5.0Å range:
|
Bromine binding site 3 out of 8 in 4xzi
Go back to
Bromine binding site 3 out
of 8 in the Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049
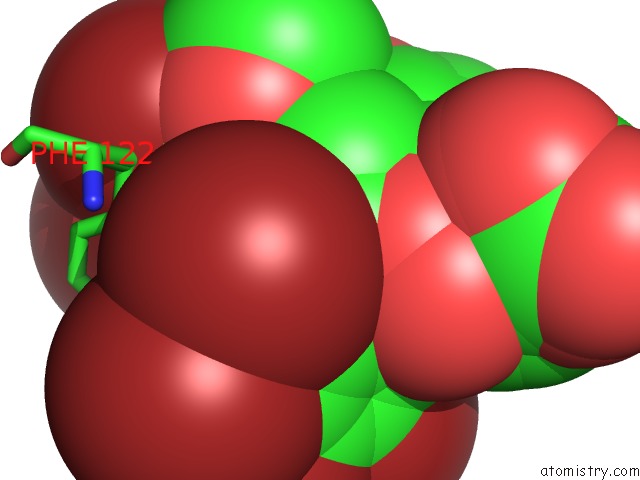
Mono view
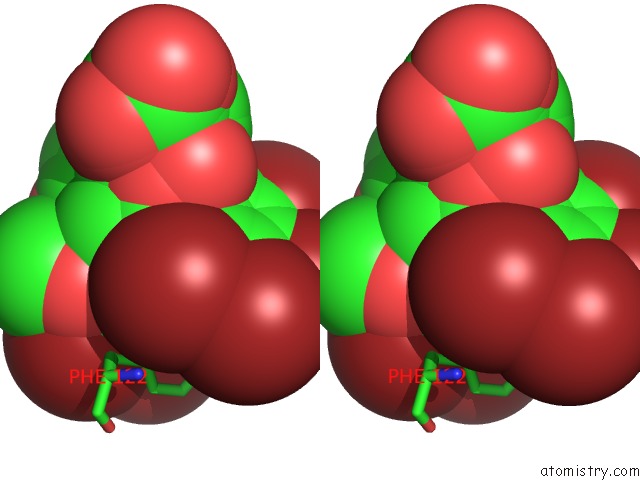
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049 within 5.0Å range:
|
Bromine binding site 4 out of 8 in 4xzi
Go back to
Bromine binding site 4 out
of 8 in the Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049
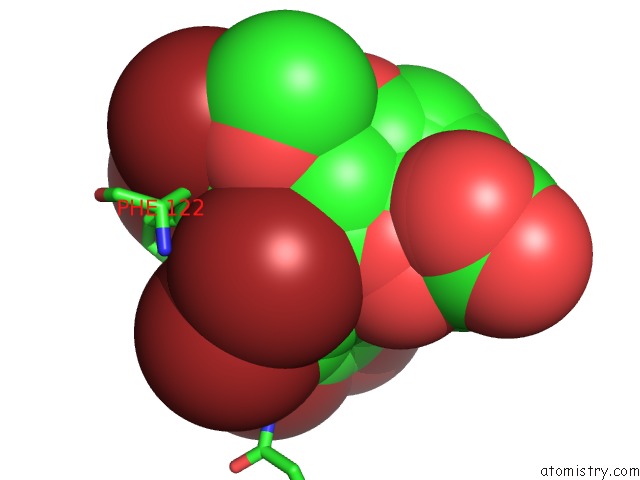
Mono view
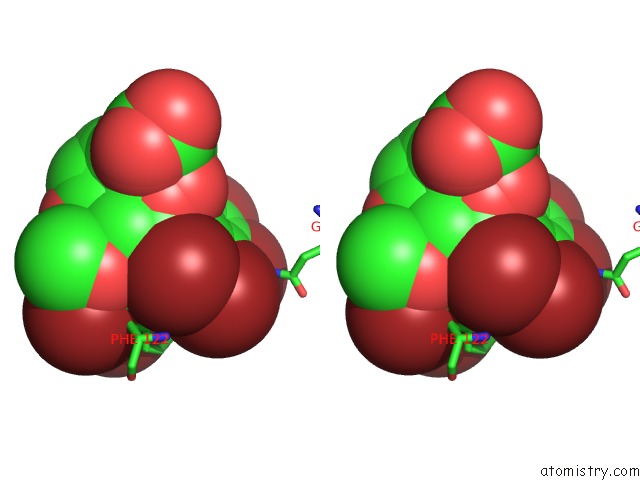
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 4 of Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049 within 5.0Å range:
|
Bromine binding site 5 out of 8 in 4xzi
Go back to
Bromine binding site 5 out
of 8 in the Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049
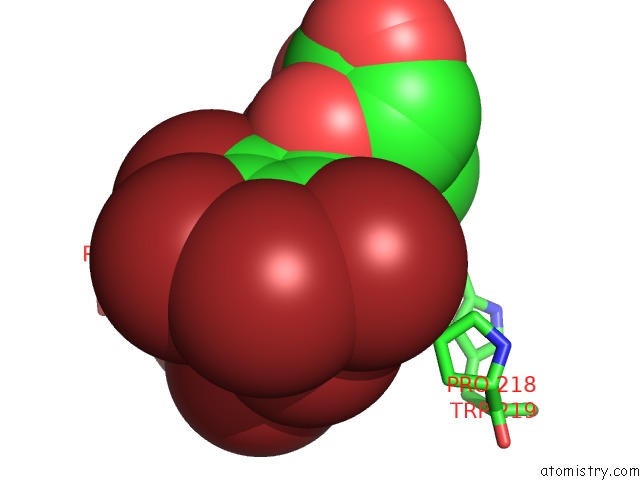
Mono view
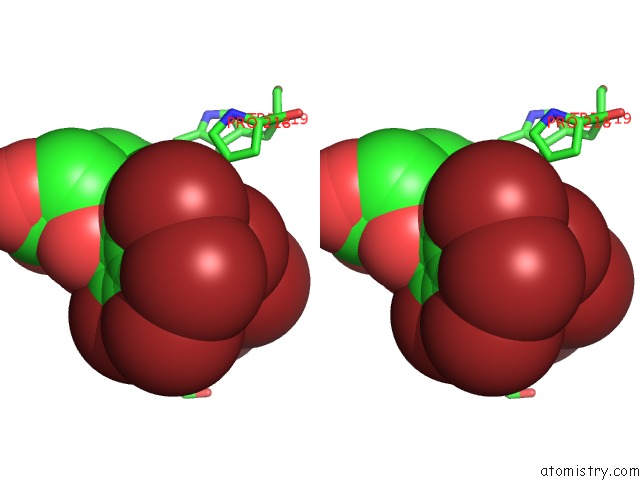
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 5 of Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049 within 5.0Å range:
|
Bromine binding site 6 out of 8 in 4xzi
Go back to
Bromine binding site 6 out
of 8 in the Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049
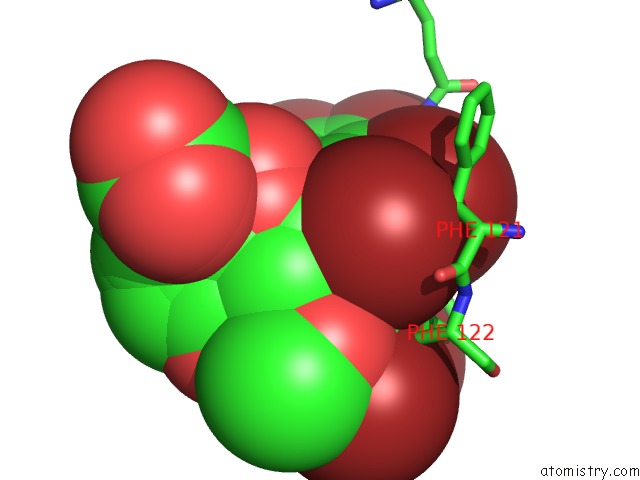
Mono view
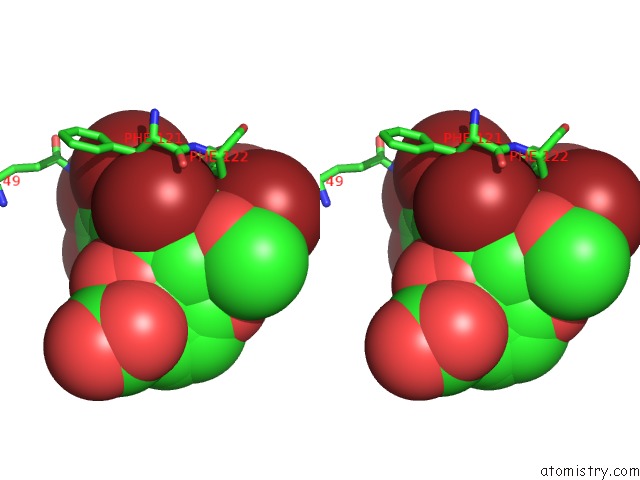
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 6 of Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049 within 5.0Å range:
|
Bromine binding site 7 out of 8 in 4xzi
Go back to
Bromine binding site 7 out
of 8 in the Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049
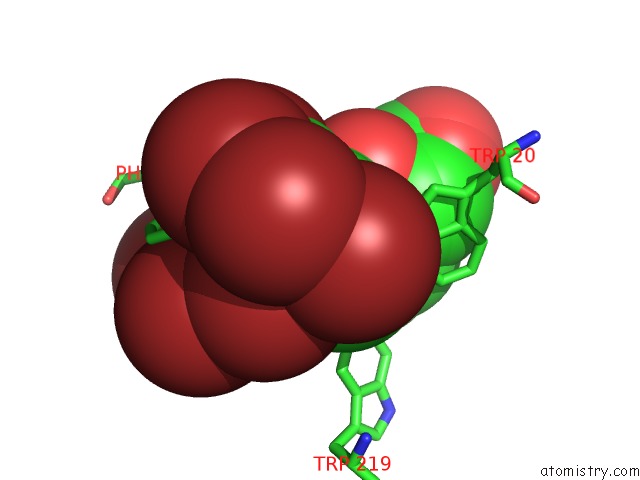
Mono view
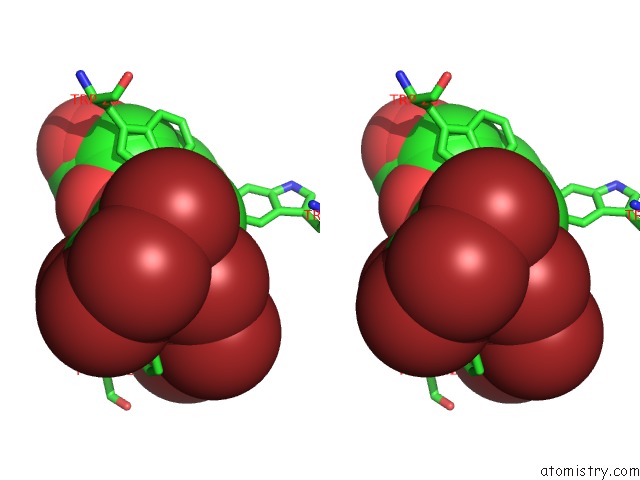
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 7 of Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049 within 5.0Å range:
|
Bromine binding site 8 out of 8 in 4xzi
Go back to
Bromine binding site 8 out
of 8 in the Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049
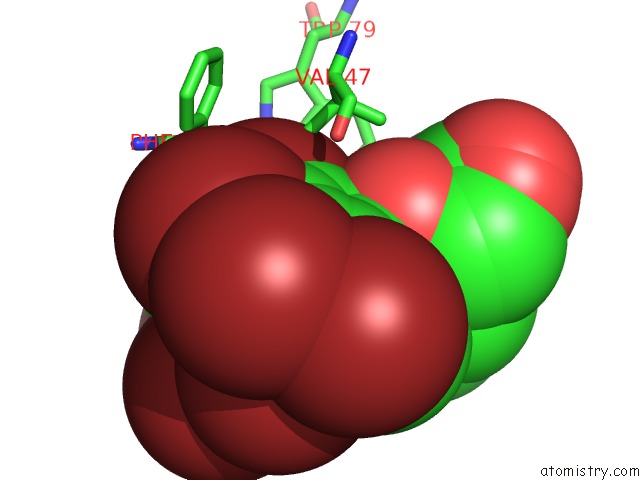
Mono view
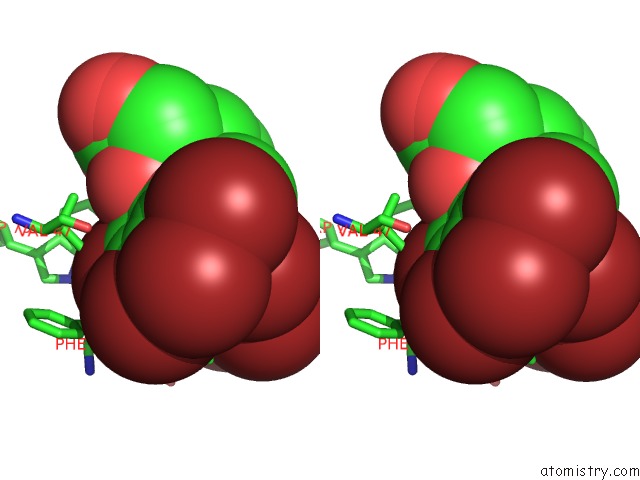
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 8 of Crystal Structure of Human Aldose Reductase Complexed with Nadp+ and JF0049 within 5.0Å range:
|
Reference:
F.X.Ruiz,
A.Cousido-Siah,
S.Porte,
M.Dominguez,
I.Crespo,
C.Rechlin,
A.Mitschler,
A.R.De Lera,
M.J.Martin,
J.A.De La Fuente,
G.Klebe,
X.Pares,
J.Farres,
A.Podjarny.
Structural Determinants of the Selectivity of 3-Benzyluracil-1-Acetic Acids Toward Human Enzymes Aldose Reductase and AKR1B10. Chemmedchem V. 10 1989 2015.
ISSN: ESSN 1860-7187
PubMed: 26549844
DOI: 10.1002/CMDC.201500393
Page generated: Mon Jul 7 07:49:48 2025
ISSN: ESSN 1860-7187
PubMed: 26549844
DOI: 10.1002/CMDC.201500393
Last articles
F in 4EHGF in 4EHE
F in 4EJN
F in 4EAR
F in 4EH9
F in 4EEV
F in 4E7P
F in 4EAD
F in 4E3N
F in 4E4X