Bromine »
PDB 5i74-5l3j »
5iex »
Bromine in PDB 5iex: Crystal Structure of (R,S)-S-{4-[(5-Bromo-4-{[(2R,3R)-2-Hydroxy-1- Methylpropyl]Oxy}- Pyrimidin-2-Yl)Amino]Phenyl}-S- Cyclopropylsulfoximide Bound to CDK2
Enzymatic activity of Crystal Structure of (R,S)-S-{4-[(5-Bromo-4-{[(2R,3R)-2-Hydroxy-1- Methylpropyl]Oxy}- Pyrimidin-2-Yl)Amino]Phenyl}-S- Cyclopropylsulfoximide Bound to CDK2
All present enzymatic activity of Crystal Structure of (R,S)-S-{4-[(5-Bromo-4-{[(2R,3R)-2-Hydroxy-1- Methylpropyl]Oxy}- Pyrimidin-2-Yl)Amino]Phenyl}-S- Cyclopropylsulfoximide Bound to CDK2:
2.7.11.22;
2.7.11.22;
Protein crystallography data
The structure of Crystal Structure of (R,S)-S-{4-[(5-Bromo-4-{[(2R,3R)-2-Hydroxy-1- Methylpropyl]Oxy}- Pyrimidin-2-Yl)Amino]Phenyl}-S- Cyclopropylsulfoximide Bound to CDK2, PDB code: 5iex
was solved by
P.Ayaz,
D.Andres,
D.A.Kwiatkowski,
C.Kolbe,
P.Lienau,
G.Siemeister,
U.Luecking,
C.M.Stegmann,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.02 / 2.03 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 53.423, 71.938, 72.554, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.3 / 25.4 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Crystal Structure of (R,S)-S-{4-[(5-Bromo-4-{[(2R,3R)-2-Hydroxy-1- Methylpropyl]Oxy}- Pyrimidin-2-Yl)Amino]Phenyl}-S- Cyclopropylsulfoximide Bound to CDK2
(pdb code 5iex). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total only one binding site of Bromine was determined in the Crystal Structure of (R,S)-S-{4-[(5-Bromo-4-{[(2R,3R)-2-Hydroxy-1- Methylpropyl]Oxy}- Pyrimidin-2-Yl)Amino]Phenyl}-S- Cyclopropylsulfoximide Bound to CDK2, PDB code: 5iex:
In total only one binding site of Bromine was determined in the Crystal Structure of (R,S)-S-{4-[(5-Bromo-4-{[(2R,3R)-2-Hydroxy-1- Methylpropyl]Oxy}- Pyrimidin-2-Yl)Amino]Phenyl}-S- Cyclopropylsulfoximide Bound to CDK2, PDB code: 5iex:
Bromine binding site 1 out of 1 in 5iex
Go back to
Bromine binding site 1 out
of 1 in the Crystal Structure of (R,S)-S-{4-[(5-Bromo-4-{[(2R,3R)-2-Hydroxy-1- Methylpropyl]Oxy}- Pyrimidin-2-Yl)Amino]Phenyl}-S- Cyclopropylsulfoximide Bound to CDK2
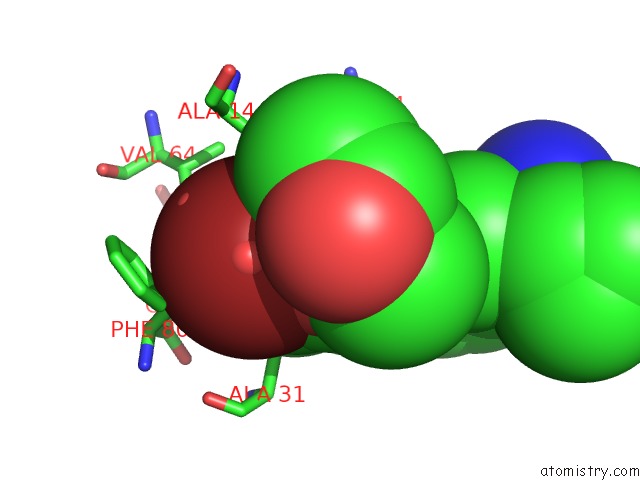
Mono view
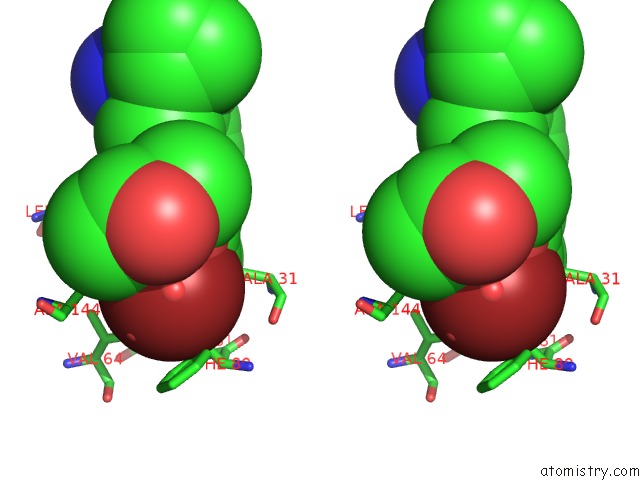
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Crystal Structure of (R,S)-S-{4-[(5-Bromo-4-{[(2R,3R)-2-Hydroxy-1- Methylpropyl]Oxy}- Pyrimidin-2-Yl)Amino]Phenyl}-S- Cyclopropylsulfoximide Bound to CDK2 within 5.0Å range:
|
Reference:
P.Ayaz,
D.Andres,
D.A.Kwiatkowski,
C.C.Kolbe,
P.Lienau,
G.Siemeister,
U.Lucking,
C.M.Stegmann.
Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (Bay 1000394), A Type I Cdk Inhibitor with Kinetic Selectivity For CDK2 and CDK9. Acs Chem.Biol. V. 11 1710 2016.
ISSN: ESSN 1554-8937
PubMed: 27090615
DOI: 10.1021/ACSCHEMBIO.6B00074
Page generated: Mon Jul 7 08:30:16 2025
ISSN: ESSN 1554-8937
PubMed: 27090615
DOI: 10.1021/ACSCHEMBIO.6B00074
Last articles
Br in 7Z97Br in 7Z77
Br in 7Z6L
Br in 7Z76
Br in 7Z57
Br in 7Z25
Br in 7Z00
Br in 7YUG
Br in 7YVX
Br in 7XKM