Bromine »
PDB 5v2h-5y93 »
5y24 »
Bromine in PDB 5y24: Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide
Protein crystallography data
The structure of Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide, PDB code: 5y24
was solved by
Q.Wang,
Z.Y.Guan,
T.T.Zou,
P.Yin,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 39.20 / 1.92 |
| Space group | P 2 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 33.550, 119.605, 214.359, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.6 / 24.9 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide
(pdb code 5y24). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 5 binding sites of Bromine where determined in the Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide, PDB code: 5y24:
Jump to Bromine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Bromine where determined in the Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide, PDB code: 5y24:
Jump to Bromine binding site number: 1; 2; 3; 4; 5;
Bromine binding site 1 out of 5 in 5y24
Go back to
Bromine binding site 1 out
of 5 in the Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide
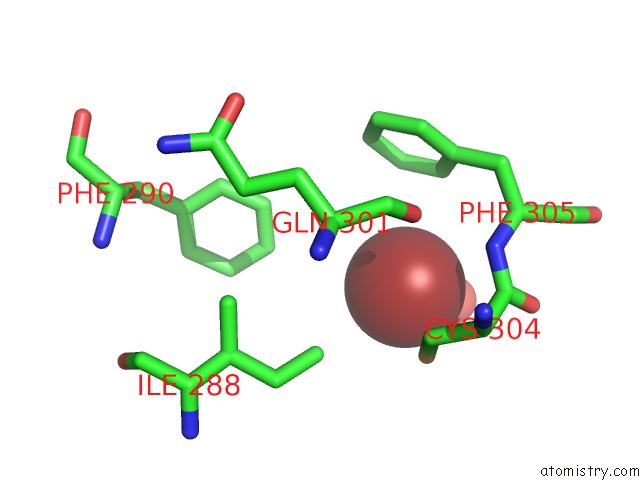
Mono view
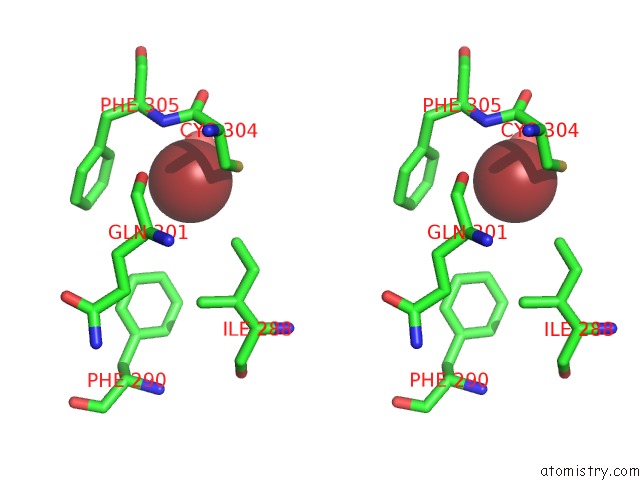
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide within 5.0Å range:
|
Bromine binding site 2 out of 5 in 5y24
Go back to
Bromine binding site 2 out
of 5 in the Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide within 5.0Å range:
|
Bromine binding site 3 out of 5 in 5y24
Go back to
Bromine binding site 3 out
of 5 in the Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide within 5.0Å range:
|
Bromine binding site 4 out of 5 in 5y24
Go back to
Bromine binding site 4 out
of 5 in the Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 4 of Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide within 5.0Å range:
|
Bromine binding site 5 out of 5 in 5y24
Go back to
Bromine binding site 5 out
of 5 in the Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 5 of Crystal Structure of Aimr From Bacillus Phage Spbeta in Complex with Its Signalling Peptide within 5.0Å range:
|
Reference:
Q.Wang,
Z.Guan,
K.Pei,
J.Wang,
Z.Liu,
P.Yin,
D.Peng,
T.Zou.
Structural Basis of the Arbitrium Peptide-Aimr Communication System in the Phage Lysis-Lysogeny Decision. Nat Microbiol V. 3 1266 2018.
ISSN: ESSN 2058-5276
PubMed: 30224798
DOI: 10.1038/S41564-018-0239-Y
Page generated: Mon Jul 7 09:27:03 2025
ISSN: ESSN 2058-5276
PubMed: 30224798
DOI: 10.1038/S41564-018-0239-Y
Last articles
Ca in 1BE8Ca in 1BE6
Ca in 1BCH
Ca in 1BCY
Ca in 1BCW
Ca in 1BCI
Ca in 1BC1
Ca in 1BC0
Ca in 1BC3
Ca in 1BAG