Bromine »
PDB 5y94-6c2x »
5yc6 »
Bromine in PDB 5yc6: The Crystal Structure of Upa in Complex with 4-Bromobenzylamirne at PH4.6
Enzymatic activity of The Crystal Structure of Upa in Complex with 4-Bromobenzylamirne at PH4.6
All present enzymatic activity of The Crystal Structure of Upa in Complex with 4-Bromobenzylamirne at PH4.6:
3.4.21.73;
3.4.21.73;
Protein crystallography data
The structure of The Crystal Structure of Upa in Complex with 4-Bromobenzylamirne at PH4.6, PDB code: 5yc6
was solved by
L.G.Jiang,
X.Zhang,
M.D.Huang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 1.18 |
| Space group | H 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 120.853, 120.853, 42.627, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 19.7 / 22.8 |
Bromine Binding Sites:
The binding sites of Bromine atom in the The Crystal Structure of Upa in Complex with 4-Bromobenzylamirne at PH4.6
(pdb code 5yc6). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total only one binding site of Bromine was determined in the The Crystal Structure of Upa in Complex with 4-Bromobenzylamirne at PH4.6, PDB code: 5yc6:
In total only one binding site of Bromine was determined in the The Crystal Structure of Upa in Complex with 4-Bromobenzylamirne at PH4.6, PDB code: 5yc6:
Bromine binding site 1 out of 1 in 5yc6
Go back to
Bromine binding site 1 out
of 1 in the The Crystal Structure of Upa in Complex with 4-Bromobenzylamirne at PH4.6
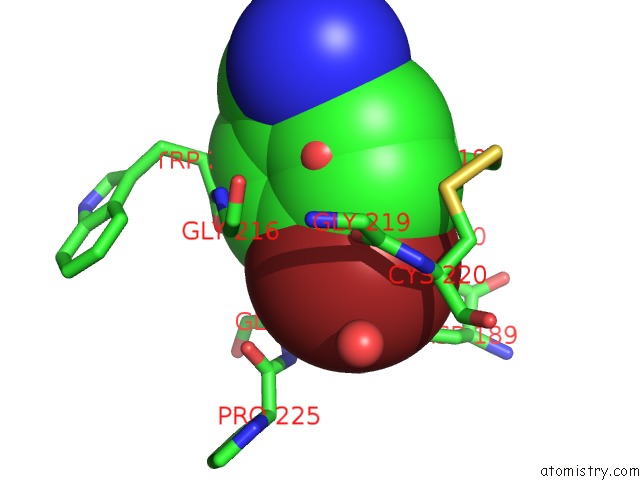
Mono view
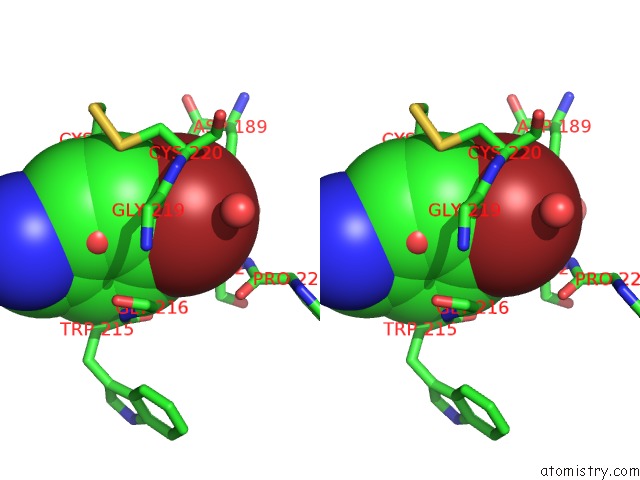
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of The Crystal Structure of Upa in Complex with 4-Bromobenzylamirne at PH4.6 within 5.0Å range:
|
Reference:
L.G.Jiang,
X.Zhang,
Y.Zhou,
Y.Y.Chen,
Z.P.Luo,
J.Y.Li,
C.Yuan,
M.D.Huang.
Halogen Bonding For the Design of Inhibitors By Targeting the S1 Pocket of Serine Proteases Rsc Adv V. 8 28189 2018.
ISSN: ESSN 2046-2069
DOI: 10.1039/C8RA03145B
Page generated: Mon Jul 7 09:28:14 2025
ISSN: ESSN 2046-2069
DOI: 10.1039/C8RA03145B
Last articles
Ca in 1CGUCa in 1CGF
Ca in 1CGT
Ca in 1CGL
Ca in 1CFF
Ca in 1CGE
Ca in 1CDM
Ca in 1CEL
Ca in 1CEH
Ca in 1C9U