Bromine »
PDB 6c3u-6e41 »
6cc9 »
Bromine in PDB 6cc9: uc(Nmr) Data-Driven Model of Gtpase Kras-Gmppnp:CMPD2 Complex Tethered to A Nanodisc
Other elements in 6cc9:
The structure of uc(Nmr) Data-Driven Model of Gtpase Kras-Gmppnp:CMPD2 Complex Tethered to A Nanodisc also contains other interesting chemical elements:
| Magnesium | (Mg) | 10 atoms |
| Chlorine | (Cl) | 10 atoms |
Bromine Binding Sites:
The binding sites of Bromine atom in the uc(Nmr) Data-Driven Model of Gtpase Kras-Gmppnp:CMPD2 Complex Tethered to A Nanodisc
(pdb code 6cc9). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total only one binding site of Bromine was determined in the uc(Nmr) Data-Driven Model of Gtpase Kras-Gmppnp:CMPD2 Complex Tethered to A Nanodisc, PDB code: 6cc9:
In total only one binding site of Bromine was determined in the uc(Nmr) Data-Driven Model of Gtpase Kras-Gmppnp:CMPD2 Complex Tethered to A Nanodisc, PDB code: 6cc9:
Bromine binding site 1 out of 1 in 6cc9
Go back to
Bromine binding site 1 out
of 1 in the uc(Nmr) Data-Driven Model of Gtpase Kras-Gmppnp:CMPD2 Complex Tethered to A Nanodisc
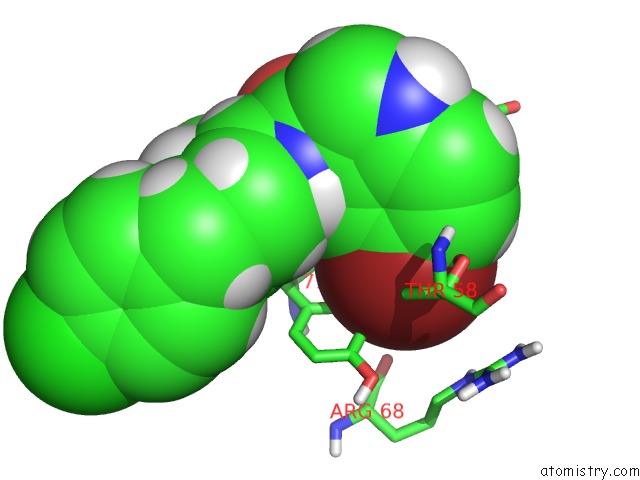
Mono view
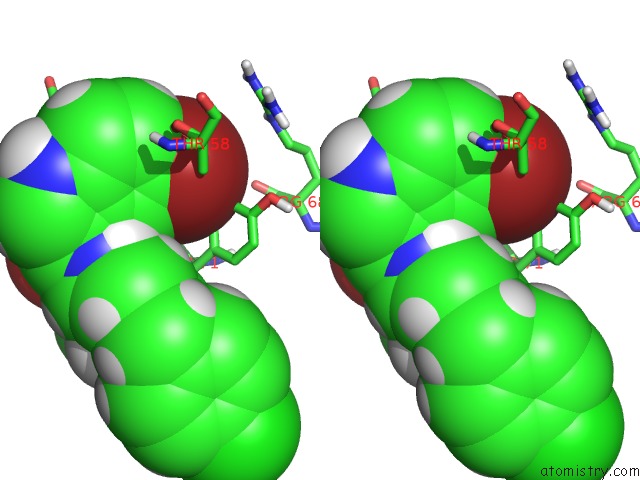
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of uc(Nmr) Data-Driven Model of Gtpase Kras-Gmppnp:CMPD2 Complex Tethered to A Nanodisc within 5.0Å range:
|
Reference:
Z.Fang,
C.B.Marshall,
T.Nishikawa,
A.D.Gossert,
J.M.Jansen,
W.Jahnke,
M.Ikura.
Inhibition of K-RAS4B By A Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site. Cell Chem Biol V. 25 1327 2018.
ISSN: ESSN 2451-9448
PubMed: 30122370
DOI: 10.1016/J.CHEMBIOL.2018.07.009
Page generated: Thu Jul 11 01:34:12 2024
ISSN: ESSN 2451-9448
PubMed: 30122370
DOI: 10.1016/J.CHEMBIOL.2018.07.009
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1