Bromine »
PDB 6hbt-6lg9 »
6ju1 »
Bromine in PDB 6ju1: P-Hydroxybenzoate Hydroxylase Y385F Mutant Complexed with 3,4- Dihydroxybenzoate
Enzymatic activity of P-Hydroxybenzoate Hydroxylase Y385F Mutant Complexed with 3,4- Dihydroxybenzoate
All present enzymatic activity of P-Hydroxybenzoate Hydroxylase Y385F Mutant Complexed with 3,4- Dihydroxybenzoate:
1.14.13.2;
1.14.13.2;
Protein crystallography data
The structure of P-Hydroxybenzoate Hydroxylase Y385F Mutant Complexed with 3,4- Dihydroxybenzoate, PDB code: 6ju1
was solved by
M.Yato,
T.Arakawa,
C.Yamada,
S.Fushinobu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.13 / 1.60 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 70.866, 145.901, 86.421, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.8 / 18.6 |
Bromine Binding Sites:
The binding sites of Bromine atom in the P-Hydroxybenzoate Hydroxylase Y385F Mutant Complexed with 3,4- Dihydroxybenzoate
(pdb code 6ju1). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 2 binding sites of Bromine where determined in the P-Hydroxybenzoate Hydroxylase Y385F Mutant Complexed with 3,4- Dihydroxybenzoate, PDB code: 6ju1:
Jump to Bromine binding site number: 1; 2;
In total 2 binding sites of Bromine where determined in the P-Hydroxybenzoate Hydroxylase Y385F Mutant Complexed with 3,4- Dihydroxybenzoate, PDB code: 6ju1:
Jump to Bromine binding site number: 1; 2;
Bromine binding site 1 out of 2 in 6ju1
Go back to
Bromine binding site 1 out
of 2 in the P-Hydroxybenzoate Hydroxylase Y385F Mutant Complexed with 3,4- Dihydroxybenzoate
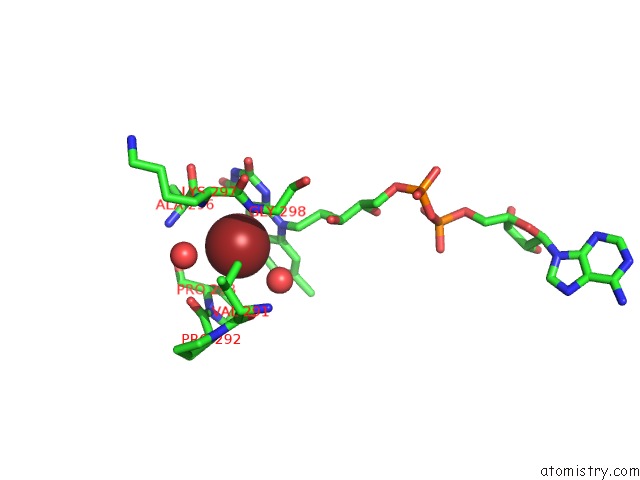
Mono view
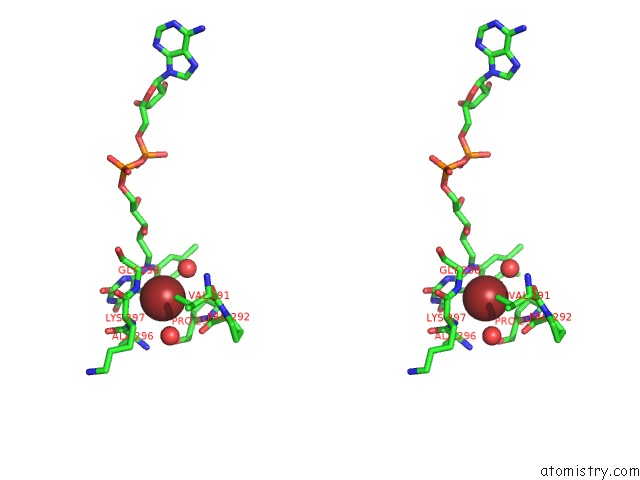
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of P-Hydroxybenzoate Hydroxylase Y385F Mutant Complexed with 3,4- Dihydroxybenzoate within 5.0Å range:
|
Bromine binding site 2 out of 2 in 6ju1
Go back to
Bromine binding site 2 out
of 2 in the P-Hydroxybenzoate Hydroxylase Y385F Mutant Complexed with 3,4- Dihydroxybenzoate
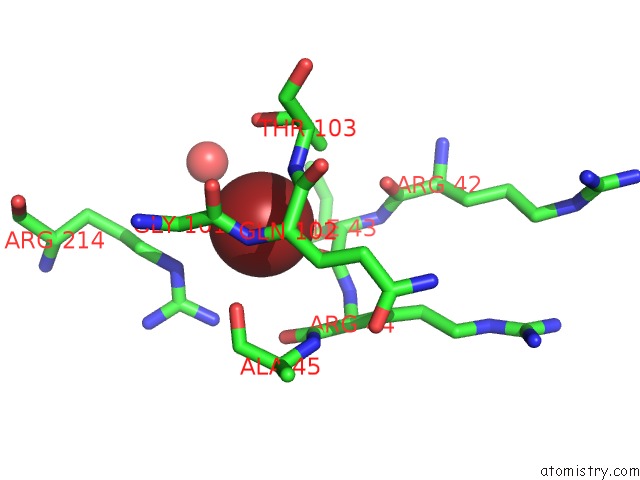
Mono view
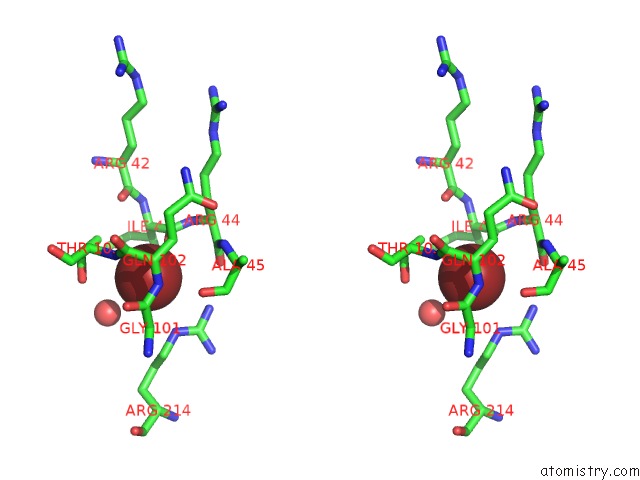
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of P-Hydroxybenzoate Hydroxylase Y385F Mutant Complexed with 3,4- Dihydroxybenzoate within 5.0Å range:
|
Reference:
Y.Moriwaki,
M.Yato,
T.Terada,
S.Saito,
N.Nukui,
T.Iwasaki,
T.Nishi,
Y.Kawaguchi,
K.Okamoto,
T.Arakawa,
C.Yamada,
S.Fushinobu,
K.Shimizu.
Understanding the Molecular Mechanism Underlying the High Catalytic Activity Ofp-Hydroxybenzoate Hydroxylase Mutants For Producing Gallic Acid. Biochemistry V. 58 4543 2019.
ISSN: ISSN 0006-2960
PubMed: 31639299
DOI: 10.1021/ACS.BIOCHEM.9B00443
Page generated: Mon Jul 7 10:04:24 2025
ISSN: ISSN 0006-2960
PubMed: 31639299
DOI: 10.1021/ACS.BIOCHEM.9B00443
Last articles
F in 7GF4F in 7GF6
F in 7GF9
F in 7GDF
F in 7GEE
F in 7GE5
F in 7GE8
F in 7GCH
F in 7GE0
F in 7GD2