Bromine »
PDB 6lue-6puu »
6npi »
Bromine in PDB 6npi: Crystal Structure of Epstein-Barr Virus Nuclear Antigen-1, EBNA1, Bound to Fragments
Protein crystallography data
The structure of Crystal Structure of Epstein-Barr Virus Nuclear Antigen-1, EBNA1, Bound to Fragments, PDB code: 6npi
was solved by
T.E.Messick,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.34 / 1.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 59.704, 67.452, 69.498, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.1 / 16.3 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Crystal Structure of Epstein-Barr Virus Nuclear Antigen-1, EBNA1, Bound to Fragments
(pdb code 6npi). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 2 binding sites of Bromine where determined in the Crystal Structure of Epstein-Barr Virus Nuclear Antigen-1, EBNA1, Bound to Fragments, PDB code: 6npi:
Jump to Bromine binding site number: 1; 2;
In total 2 binding sites of Bromine where determined in the Crystal Structure of Epstein-Barr Virus Nuclear Antigen-1, EBNA1, Bound to Fragments, PDB code: 6npi:
Jump to Bromine binding site number: 1; 2;
Bromine binding site 1 out of 2 in 6npi
Go back to
Bromine binding site 1 out
of 2 in the Crystal Structure of Epstein-Barr Virus Nuclear Antigen-1, EBNA1, Bound to Fragments
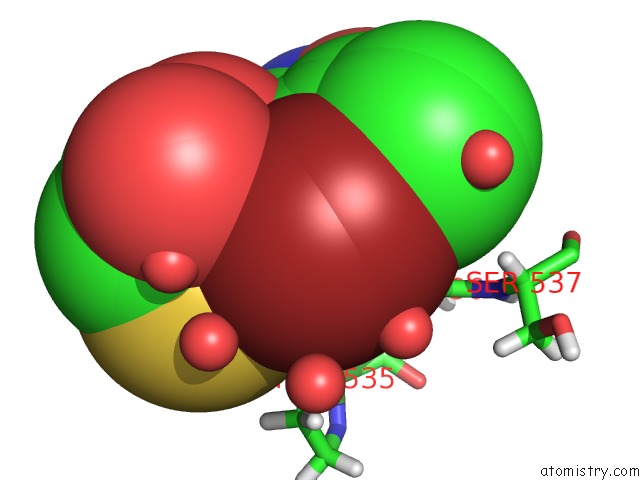
Mono view
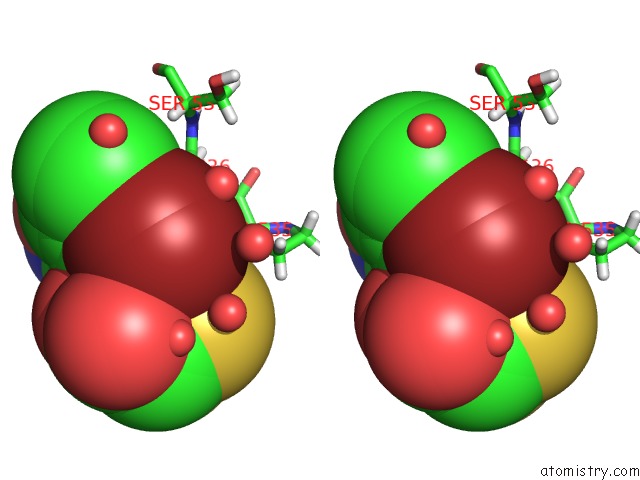
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Crystal Structure of Epstein-Barr Virus Nuclear Antigen-1, EBNA1, Bound to Fragments within 5.0Å range:
|
Bromine binding site 2 out of 2 in 6npi
Go back to
Bromine binding site 2 out
of 2 in the Crystal Structure of Epstein-Barr Virus Nuclear Antigen-1, EBNA1, Bound to Fragments
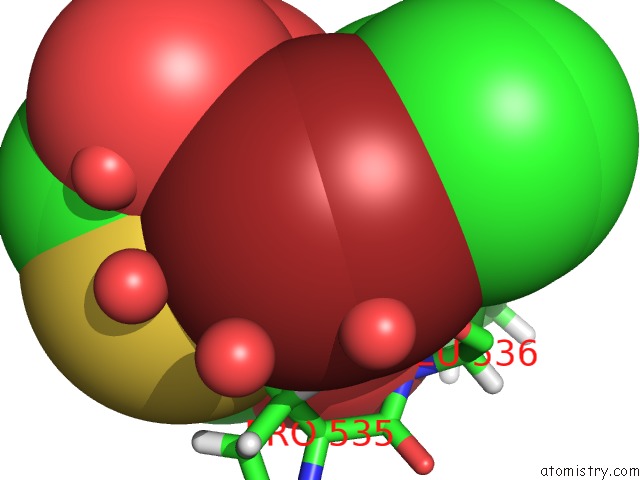
Mono view
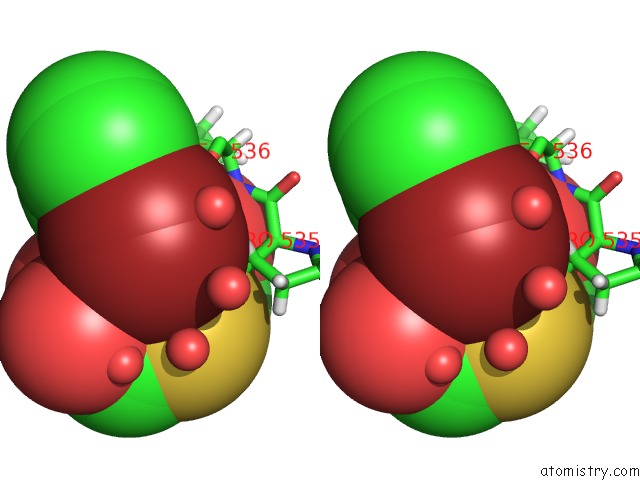
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Crystal Structure of Epstein-Barr Virus Nuclear Antigen-1, EBNA1, Bound to Fragments within 5.0Å range:
|
Reference:
T.E.Messick,
G.R.Smith,
S.S.Soldan,
M.E.Mcdonnell,
J.S.Deakyne,
K.A.Malecka,
L.Tolvinski,
A.P.J.Van Den Heuvel,
B.W.Gu,
J.A.Cassel,
D.H.Tran,
B.R.Wassermann,
Y.Zhang,
V.Velvadapu,
E.R.Zartler,
P.Busson,
A.B.Reitz,
P.M.Lieberman.
Structure-Based Design of Small-Molecule Inhibitors of EBNA1 Dna Binding Blocks Epstein-Barr Virus Latent Infection and Tumor Growth. Sci Transl Med V. 11 2019.
ISSN: ESSN 1946-6242
PubMed: 30842315
DOI: 10.1126/SCITRANSLMED.AAU5612
Page generated: Mon Jul 7 10:09:25 2025
ISSN: ESSN 1946-6242
PubMed: 30842315
DOI: 10.1126/SCITRANSLMED.AAU5612
Last articles
Cl in 3KXRCl in 3KWE
Cl in 3KWK
Cl in 3KWD
Cl in 3KWC
Cl in 3KW4
Cl in 3KVW
Cl in 3KUR
Cl in 3KVK
Cl in 3KVM