Bromine »
PDB 6tu4-6vrk »
6ujp »
Bromine in PDB 6ujp: P-Glycoprotein Mutant-F979A and C952A-with BDE100
Enzymatic activity of P-Glycoprotein Mutant-F979A and C952A-with BDE100
All present enzymatic activity of P-Glycoprotein Mutant-F979A and C952A-with BDE100:
7.6.2.1; 7.6.2.2;
7.6.2.1; 7.6.2.2;
Protein crystallography data
The structure of P-Glycoprotein Mutant-F979A and C952A-with BDE100, PDB code: 6ujp
was solved by
S.G.Aller,
C.A.Le,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.59 / 3.98 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 89.046, 138.133, 188.246, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 23.9 / 26.3 |
Bromine Binding Sites:
The binding sites of Bromine atom in the P-Glycoprotein Mutant-F979A and C952A-with BDE100
(pdb code 6ujp). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 5 binding sites of Bromine where determined in the P-Glycoprotein Mutant-F979A and C952A-with BDE100, PDB code: 6ujp:
Jump to Bromine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Bromine where determined in the P-Glycoprotein Mutant-F979A and C952A-with BDE100, PDB code: 6ujp:
Jump to Bromine binding site number: 1; 2; 3; 4; 5;
Bromine binding site 1 out of 5 in 6ujp
Go back to
Bromine binding site 1 out
of 5 in the P-Glycoprotein Mutant-F979A and C952A-with BDE100
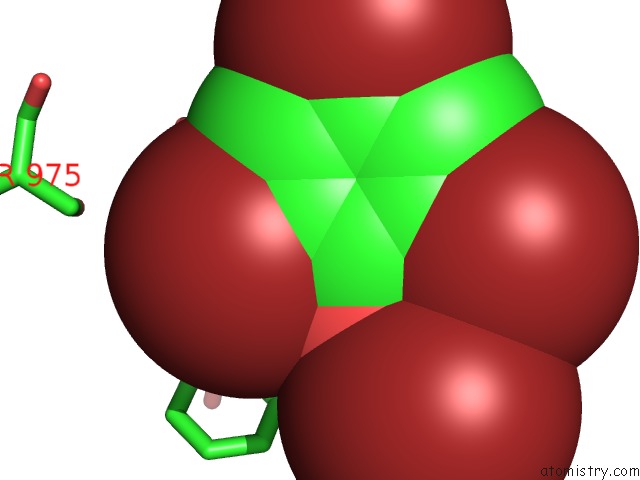
Mono view
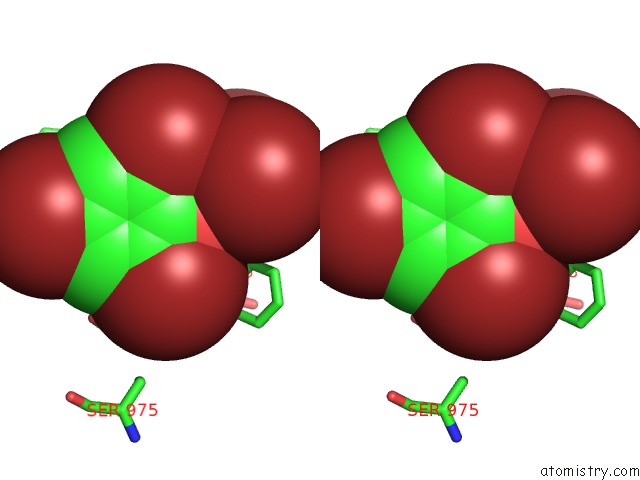
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of P-Glycoprotein Mutant-F979A and C952A-with BDE100 within 5.0Å range:
|
Bromine binding site 2 out of 5 in 6ujp
Go back to
Bromine binding site 2 out
of 5 in the P-Glycoprotein Mutant-F979A and C952A-with BDE100
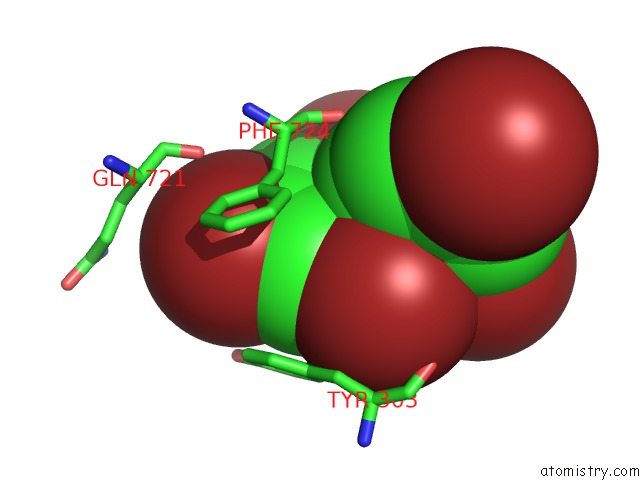
Mono view
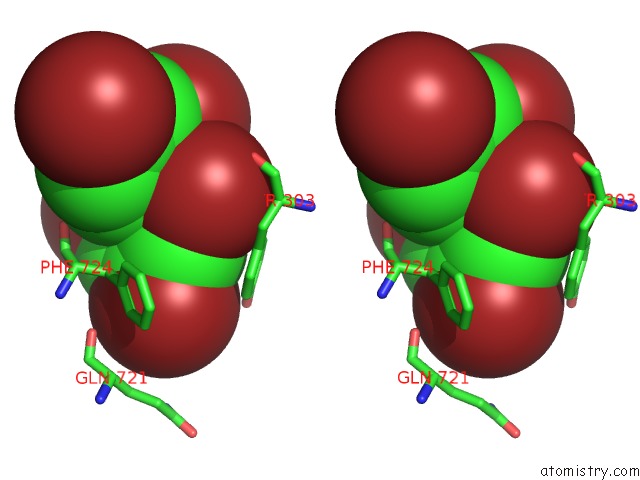
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of P-Glycoprotein Mutant-F979A and C952A-with BDE100 within 5.0Å range:
|
Bromine binding site 3 out of 5 in 6ujp
Go back to
Bromine binding site 3 out
of 5 in the P-Glycoprotein Mutant-F979A and C952A-with BDE100
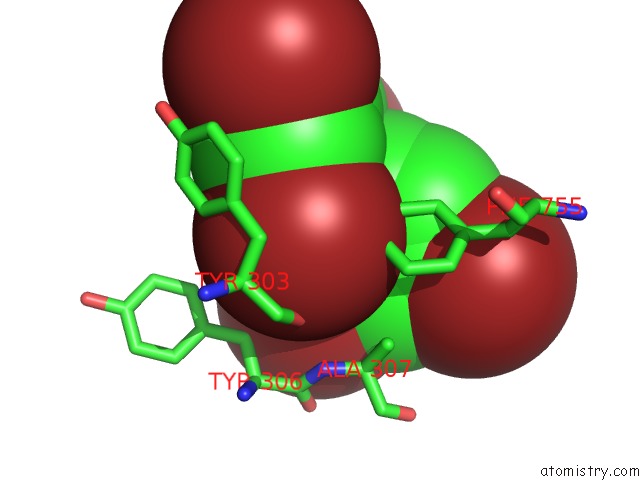
Mono view
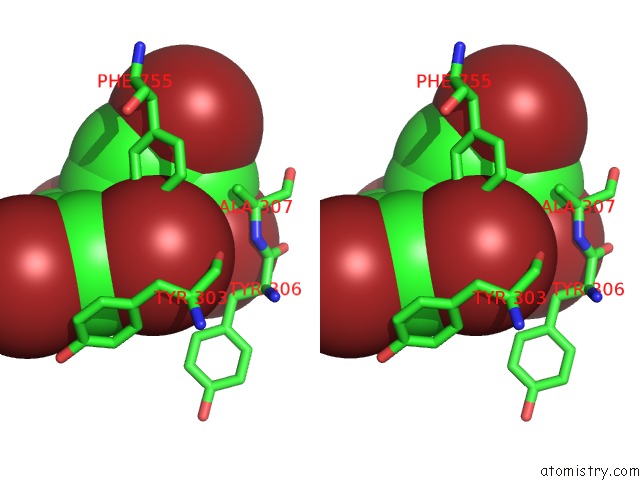
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of P-Glycoprotein Mutant-F979A and C952A-with BDE100 within 5.0Å range:
|
Bromine binding site 4 out of 5 in 6ujp
Go back to
Bromine binding site 4 out
of 5 in the P-Glycoprotein Mutant-F979A and C952A-with BDE100
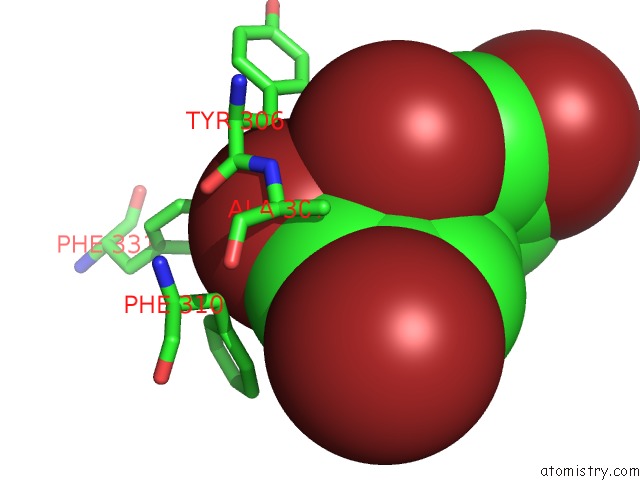
Mono view
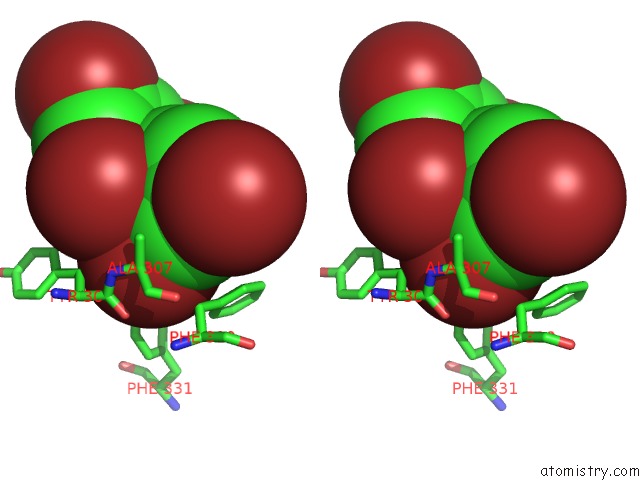
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 4 of P-Glycoprotein Mutant-F979A and C952A-with BDE100 within 5.0Å range:
|
Bromine binding site 5 out of 5 in 6ujp
Go back to
Bromine binding site 5 out
of 5 in the P-Glycoprotein Mutant-F979A and C952A-with BDE100
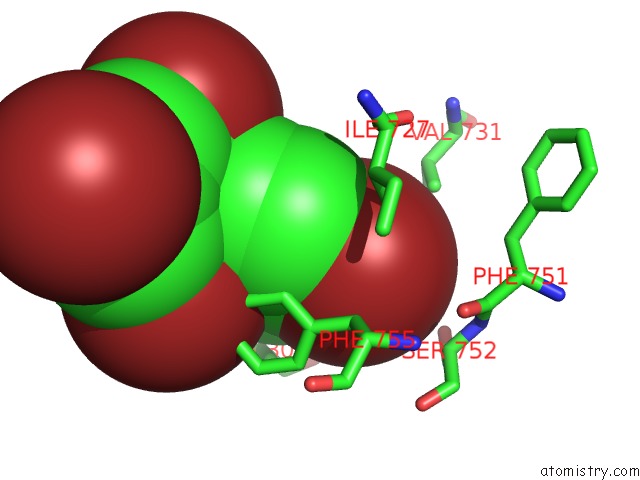
Mono view
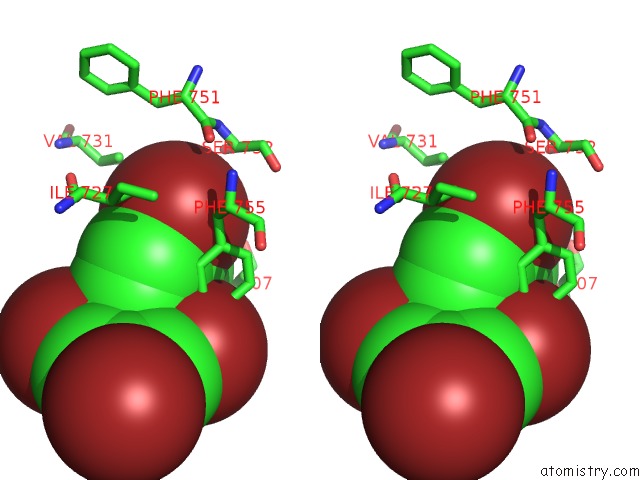
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 5 of P-Glycoprotein Mutant-F979A and C952A-with BDE100 within 5.0Å range:
|
Reference:
C.A.Le,
D.S.Harvey,
S.G.Aller.
Structural Definition of Polyspecific Compensatory Ligand Recognition By Pglycoprotein Iucrj 2020.
ISSN: ESSN 2052-2525
DOI: 10.1107/S2052252520005709
Page generated: Mon Jul 7 10:34:51 2025
ISSN: ESSN 2052-2525
DOI: 10.1107/S2052252520005709
Last articles
Cl in 7XGNCl in 7XHH
Cl in 7XGM
Cl in 7XHK
Cl in 7XGL
Cl in 7XGI
Cl in 7XFY
Cl in 7XFA
Cl in 7XFX
Cl in 7XFW