Bromine »
PDB 7qwb-7u5q »
7rde »
Bromine in PDB 7rde: Human Triose Phosphate Isomerase Q181P
Enzymatic activity of Human Triose Phosphate Isomerase Q181P
Protein crystallography data
The structure of Human Triose Phosphate Isomerase Q181P, PDB code: 7rde
was solved by
A.P.Vandemark,
J.Kowalski,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.33 / 1.31 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 73.189, 77.822, 84.295, 90, 90, 90 |
| R / Rfree (%) | 13.7 / 16.2 |
Other elements in 7rde:
The structure of Human Triose Phosphate Isomerase Q181P also contains other interesting chemical elements:
| Calcium | (Ca) | 1 atom |
Bromine Binding Sites:
The binding sites of Bromine atom in the Human Triose Phosphate Isomerase Q181P
(pdb code 7rde). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 2 binding sites of Bromine where determined in the Human Triose Phosphate Isomerase Q181P, PDB code: 7rde:
Jump to Bromine binding site number: 1; 2;
In total 2 binding sites of Bromine where determined in the Human Triose Phosphate Isomerase Q181P, PDB code: 7rde:
Jump to Bromine binding site number: 1; 2;
Bromine binding site 1 out of 2 in 7rde
Go back to
Bromine binding site 1 out
of 2 in the Human Triose Phosphate Isomerase Q181P

Mono view
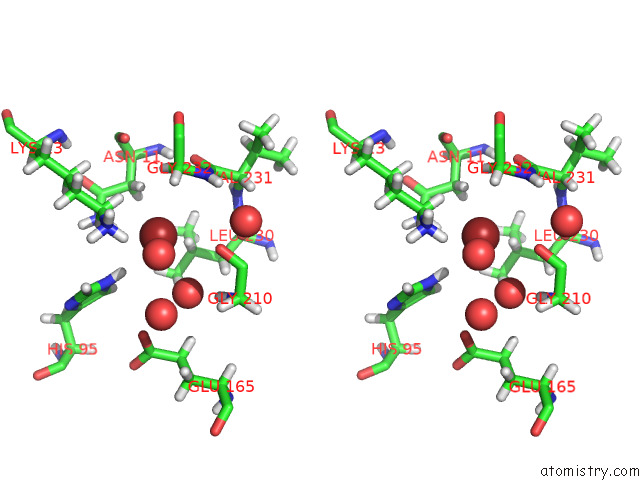
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Human Triose Phosphate Isomerase Q181P within 5.0Å range:
|
Bromine binding site 2 out of 2 in 7rde
Go back to
Bromine binding site 2 out
of 2 in the Human Triose Phosphate Isomerase Q181P

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Human Triose Phosphate Isomerase Q181P within 5.0Å range:
|
Reference:
A.P.Vandemark,
S.L.Hrizo,
S.L.Eicher,
J.Kowalski,
T.D.Myers,
M.R.Pfeifer,
K.N.Riley,
D.D.Koeberl,
M.J.Palladino.
Itavastatin and Resveratrol Increase Triosephosphate Isomerase Protein in A Newly Identified Variant of Tpi Deficiency. Dis Model Mech V. 15 2022.
ISSN: ISSN 1754-8411
PubMed: 35315486
DOI: 10.1242/DMM.049261
Page generated: Mon Jul 7 11:46:32 2025
ISSN: ISSN 1754-8411
PubMed: 35315486
DOI: 10.1242/DMM.049261
Last articles
Ca in 5M0SCa in 5LYJ
Ca in 5LXV
Ca in 5M0M
Ca in 5M0E
Ca in 5M0D
Ca in 5LZH
Ca in 5M06
Ca in 5LXT
Ca in 5LXS