Bromine »
PDB 7uc6-7yc9 »
7vso »
Bromine in PDB 7vso: Serial Femtosecond Crystallography (Sfx) of Ground State Bacteriorhodopsin Crystallized From Bicelles in Complex with HAD16 Determined Using 7-Kev X-Ray Free Electron Laser (Xfel) at Sacla
Protein crystallography data
The structure of Serial Femtosecond Crystallography (Sfx) of Ground State Bacteriorhodopsin Crystallized From Bicelles in Complex with HAD16 Determined Using 7-Kev X-Ray Free Electron Laser (Xfel) at Sacla, PDB code: 7vso
was solved by
E.Mizohata,
T.Nakane,
S.Hanashima,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.90 / 2.35 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 46.2, 103, 128.7, 90, 90, 90 |
| R / Rfree (%) | 15.8 / 21 |
Other elements in 7vso:
The structure of Serial Femtosecond Crystallography (Sfx) of Ground State Bacteriorhodopsin Crystallized From Bicelles in Complex with HAD16 Determined Using 7-Kev X-Ray Free Electron Laser (Xfel) at Sacla also contains other interesting chemical elements:
| Iodine | (I) | 3 atoms |
Bromine Binding Sites:
The binding sites of Bromine atom in the Serial Femtosecond Crystallography (Sfx) of Ground State Bacteriorhodopsin Crystallized From Bicelles in Complex with HAD16 Determined Using 7-Kev X-Ray Free Electron Laser (Xfel) at Sacla
(pdb code 7vso). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 2 binding sites of Bromine where determined in the Serial Femtosecond Crystallography (Sfx) of Ground State Bacteriorhodopsin Crystallized From Bicelles in Complex with HAD16 Determined Using 7-Kev X-Ray Free Electron Laser (Xfel) at Sacla, PDB code: 7vso:
Jump to Bromine binding site number: 1; 2;
In total 2 binding sites of Bromine where determined in the Serial Femtosecond Crystallography (Sfx) of Ground State Bacteriorhodopsin Crystallized From Bicelles in Complex with HAD16 Determined Using 7-Kev X-Ray Free Electron Laser (Xfel) at Sacla, PDB code: 7vso:
Jump to Bromine binding site number: 1; 2;
Bromine binding site 1 out of 2 in 7vso
Go back to
Bromine binding site 1 out
of 2 in the Serial Femtosecond Crystallography (Sfx) of Ground State Bacteriorhodopsin Crystallized From Bicelles in Complex with HAD16 Determined Using 7-Kev X-Ray Free Electron Laser (Xfel) at Sacla
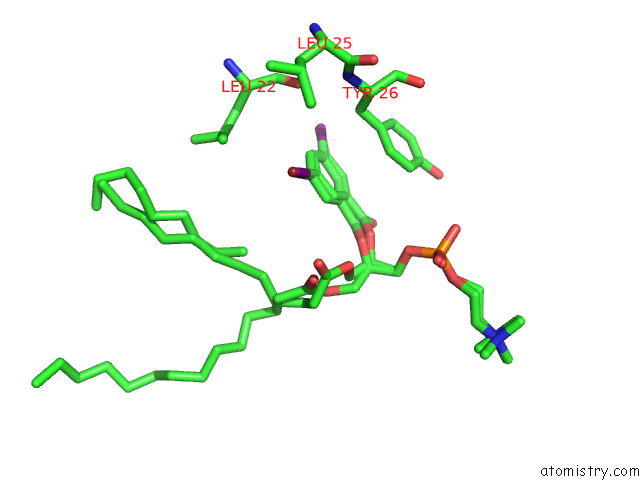
Mono view
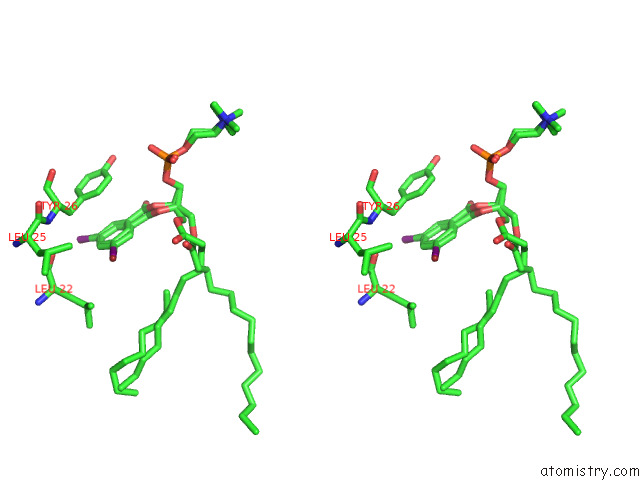
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Serial Femtosecond Crystallography (Sfx) of Ground State Bacteriorhodopsin Crystallized From Bicelles in Complex with HAD16 Determined Using 7-Kev X-Ray Free Electron Laser (Xfel) at Sacla within 5.0Å range:
|
Bromine binding site 2 out of 2 in 7vso
Go back to
Bromine binding site 2 out
of 2 in the Serial Femtosecond Crystallography (Sfx) of Ground State Bacteriorhodopsin Crystallized From Bicelles in Complex with HAD16 Determined Using 7-Kev X-Ray Free Electron Laser (Xfel) at Sacla
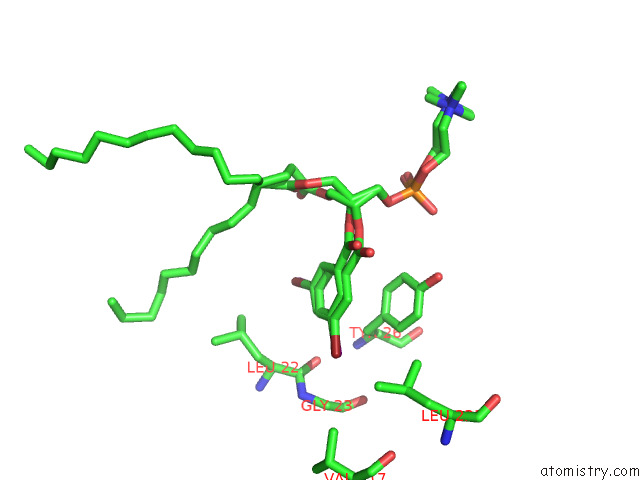
Mono view
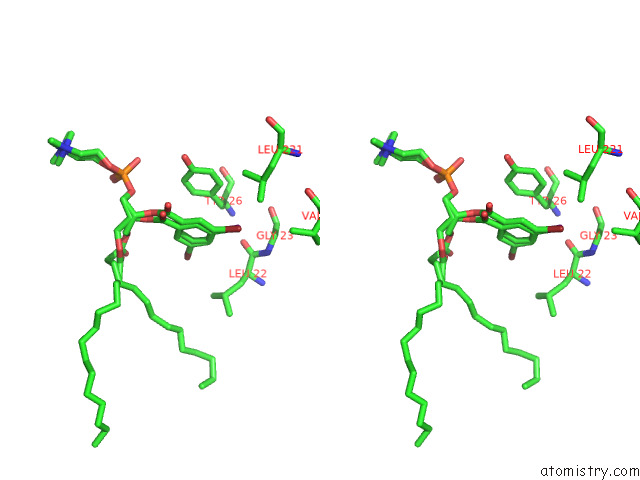
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Serial Femtosecond Crystallography (Sfx) of Ground State Bacteriorhodopsin Crystallized From Bicelles in Complex with HAD16 Determined Using 7-Kev X-Ray Free Electron Laser (Xfel) at Sacla within 5.0Å range:
|
Reference:
S.Hanashima,
T.Nakane,
E.Mizohata.
Heavy Atom Detergent/Lipid Combined X-Ray Crystallography For Elucidating the Structure-Function Relationships of Membrane Proteins. Membranes (Basel) V. 11 2021.
ISSN: ESSN 2077-0375
PubMed: 34832053
DOI: 10.3390/MEMBRANES11110823
Page generated: Mon Jul 7 11:53:23 2025
ISSN: ESSN 2077-0375
PubMed: 34832053
DOI: 10.3390/MEMBRANES11110823
Last articles
Fe in 2EUUFe in 2EUT
Fe in 2EVP
Fe in 2EVK
Fe in 2EUS
Fe in 2EUR
Fe in 2E84
Fe in 2EUQ
Fe in 2ET0
Fe in 2EUP