Bromine »
PDB 8adq-8d90 »
8d87 »
Bromine in PDB 8d87: Fitted Crystal Structure of the Homotrimer of Fusion Glycoprotein E1 From Sfv Into Subtomogram Averaged Chikv E1 Glycoprotein Density
Other elements in 8d87:
The structure of Fitted Crystal Structure of the Homotrimer of Fusion Glycoprotein E1 From Sfv Into Subtomogram Averaged Chikv E1 Glycoprotein Density also contains other interesting chemical elements:
| Holmium | (Ho) | 4 atoms |
Bromine Binding Sites:
The binding sites of Bromine atom in the Fitted Crystal Structure of the Homotrimer of Fusion Glycoprotein E1 From Sfv Into Subtomogram Averaged Chikv E1 Glycoprotein Density
(pdb code 8d87). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 3 binding sites of Bromine where determined in the Fitted Crystal Structure of the Homotrimer of Fusion Glycoprotein E1 From Sfv Into Subtomogram Averaged Chikv E1 Glycoprotein Density, PDB code: 8d87:
Jump to Bromine binding site number: 1; 2; 3;
In total 3 binding sites of Bromine where determined in the Fitted Crystal Structure of the Homotrimer of Fusion Glycoprotein E1 From Sfv Into Subtomogram Averaged Chikv E1 Glycoprotein Density, PDB code: 8d87:
Jump to Bromine binding site number: 1; 2; 3;
Bromine binding site 1 out of 3 in 8d87
Go back to
Bromine binding site 1 out
of 3 in the Fitted Crystal Structure of the Homotrimer of Fusion Glycoprotein E1 From Sfv Into Subtomogram Averaged Chikv E1 Glycoprotein Density
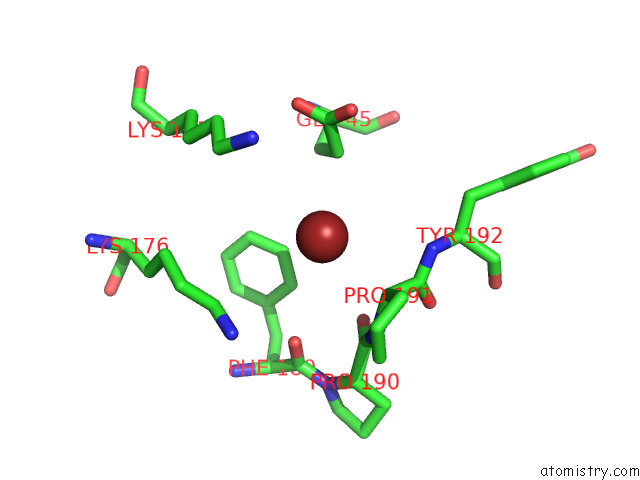
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Fitted Crystal Structure of the Homotrimer of Fusion Glycoprotein E1 From Sfv Into Subtomogram Averaged Chikv E1 Glycoprotein Density within 5.0Å range:
|
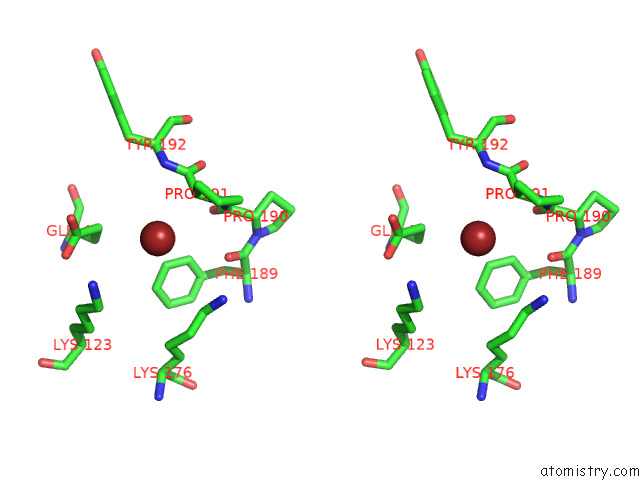
Bromine binding site 2 out of 3 in 8d87
Go back to
Bromine binding site 2 out
of 3 in the Fitted Crystal Structure of the Homotrimer of Fusion Glycoprotein E1 From Sfv Into Subtomogram Averaged Chikv E1 Glycoprotein Density
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Fitted Crystal Structure of the Homotrimer of Fusion Glycoprotein E1 From Sfv Into Subtomogram Averaged Chikv E1 Glycoprotein Density within 5.0Å range:
|
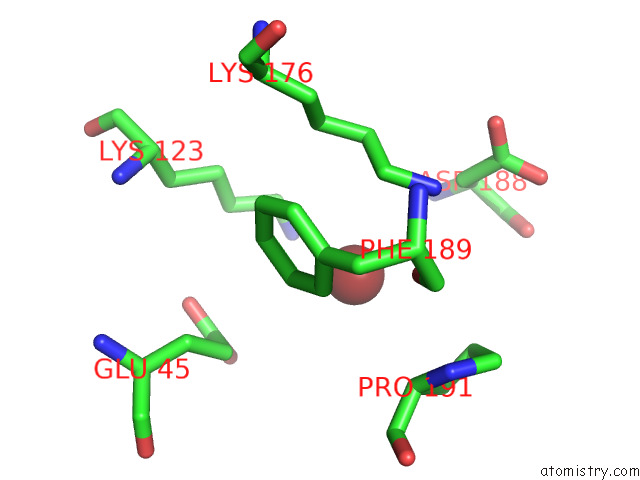
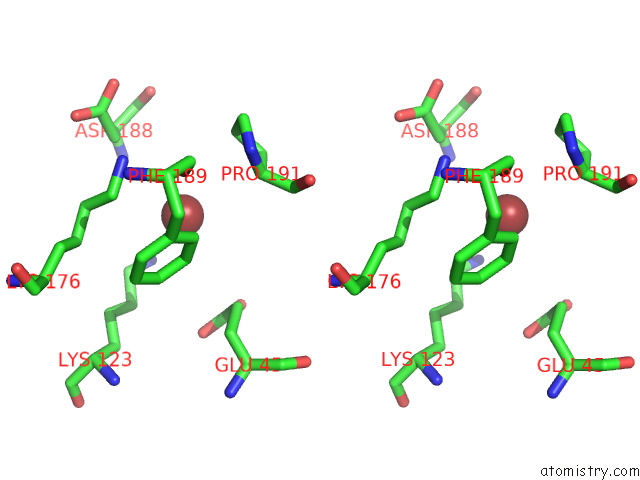
Bromine binding site 3 out of 3 in 8d87
Go back to
Bromine binding site 3 out
of 3 in the Fitted Crystal Structure of the Homotrimer of Fusion Glycoprotein E1 From Sfv Into Subtomogram Averaged Chikv E1 Glycoprotein Density
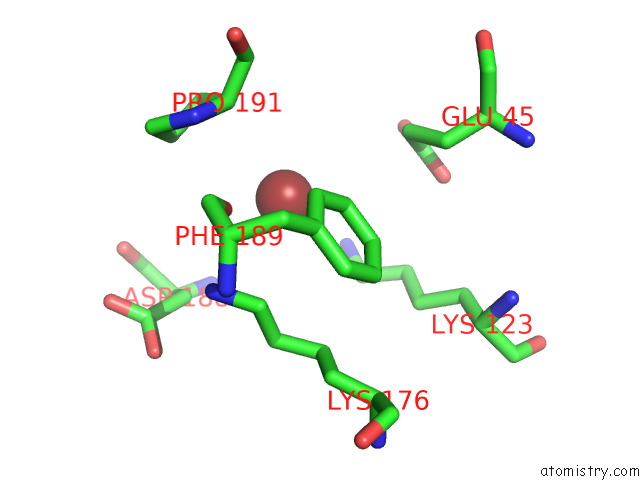
Mono view
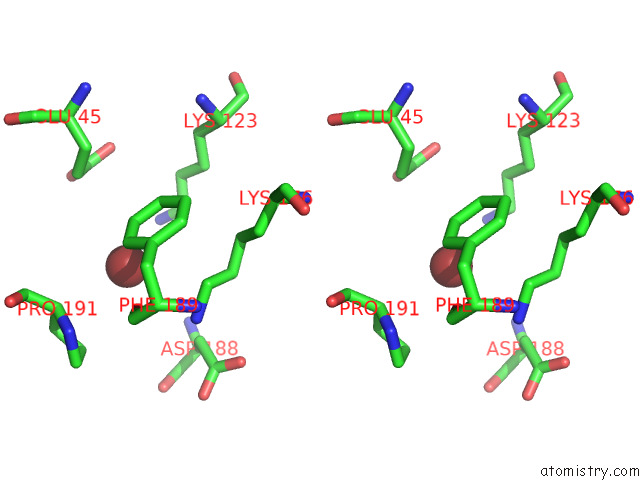
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of Fitted Crystal Structure of the Homotrimer of Fusion Glycoprotein E1 From Sfv Into Subtomogram Averaged Chikv E1 Glycoprotein Density within 5.0Å range:
|
Reference:
V.Mangala Prasad,
J.S.Blijleven,
J.M.Smit,
K.K.Lee.
Visualization of Conformational Changes and Membrane Remodeling Leading to Genome Delivery By Viral Class-II Fusion Machinery. Nat Commun V. 13 4772 2022.
ISSN: ESSN 2041-1723
PubMed: 35970990
DOI: 10.1038/S41467-022-32431-9
Page generated: Thu Jul 11 05:01:05 2024
ISSN: ESSN 2041-1723
PubMed: 35970990
DOI: 10.1038/S41467-022-32431-9
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1