Bromine »
PDB 8d90-8gb1 »
8dq0 »
Bromine in PDB 8dq0: Quorum-Sensing Receptor Rhlr Bound to Pqse
Enzymatic activity of Quorum-Sensing Receptor Rhlr Bound to Pqse
All present enzymatic activity of Quorum-Sensing Receptor Rhlr Bound to Pqse:
3.1.2.32;
3.1.2.32;
Bromine Binding Sites:
The binding sites of Bromine atom in the Quorum-Sensing Receptor Rhlr Bound to Pqse
(pdb code 8dq0). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 2 binding sites of Bromine where determined in the Quorum-Sensing Receptor Rhlr Bound to Pqse, PDB code: 8dq0:
Jump to Bromine binding site number: 1; 2;
In total 2 binding sites of Bromine where determined in the Quorum-Sensing Receptor Rhlr Bound to Pqse, PDB code: 8dq0:
Jump to Bromine binding site number: 1; 2;
Bromine binding site 1 out of 2 in 8dq0
Go back to
Bromine binding site 1 out
of 2 in the Quorum-Sensing Receptor Rhlr Bound to Pqse
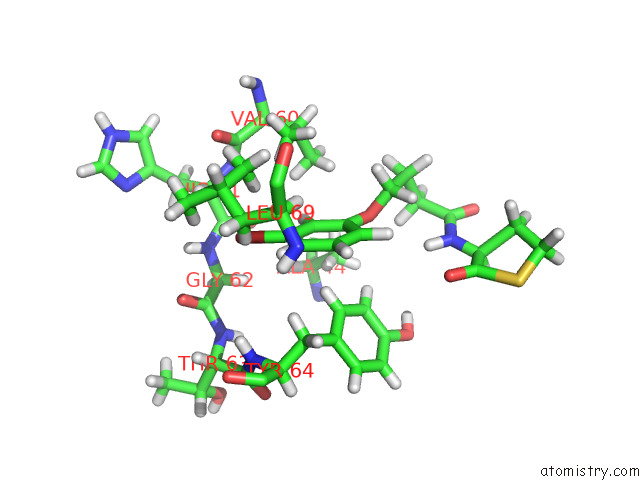
Mono view
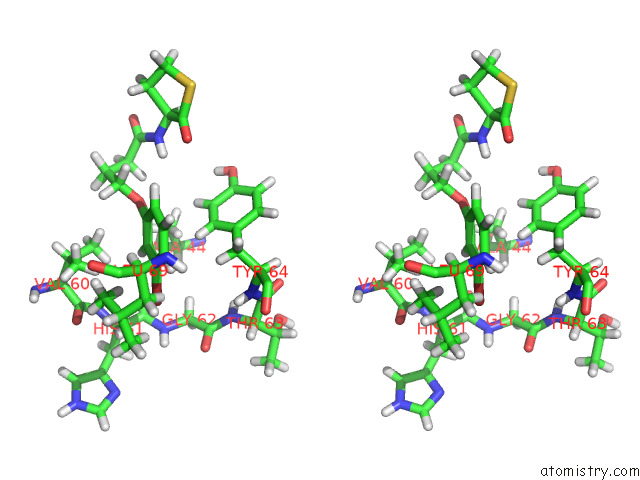
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Quorum-Sensing Receptor Rhlr Bound to Pqse within 5.0Å range:
|
Bromine binding site 2 out of 2 in 8dq0
Go back to
Bromine binding site 2 out
of 2 in the Quorum-Sensing Receptor Rhlr Bound to Pqse
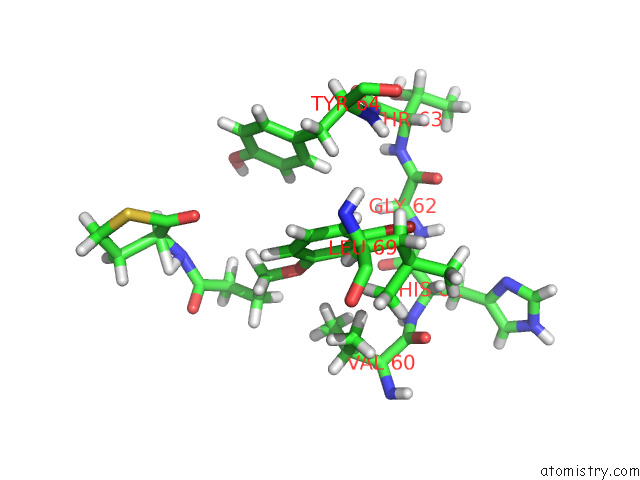
Mono view
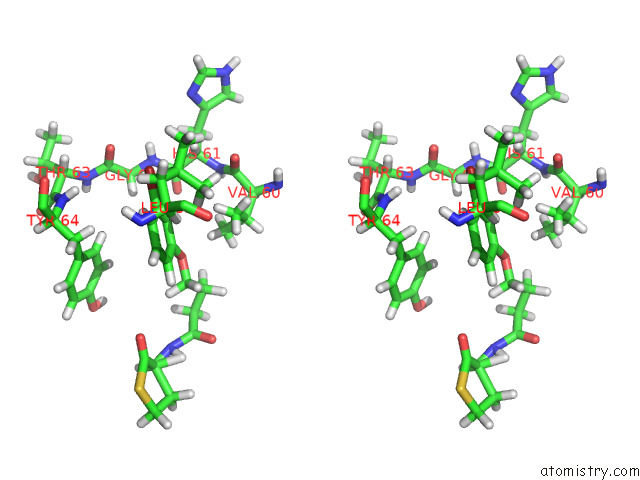
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Quorum-Sensing Receptor Rhlr Bound to Pqse within 5.0Å range:
|
Reference:
J.R.Feathers,
E.K.Richael,
K.A.Simanek,
J.C.Fromme,
J.E.Paczkowski.
Structure of the Rhlr-Pqse Complex From Pseudomonas Aeruginosa Reveals Mechanistic Insights Into Quorum-Sensing Gene Regulation. Structure V. 30 1626 2022.
ISSN: ISSN 0969-2126
PubMed: 36379213
DOI: 10.1016/J.STR.2022.10.008
Page generated: Mon Jul 7 12:11:52 2025
ISSN: ISSN 0969-2126
PubMed: 36379213
DOI: 10.1016/J.STR.2022.10.008
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO