Bromine »
PDB 8d90-8gb1 »
8g0c »
Bromine in PDB 8g0c: Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model)
Enzymatic activity of Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model)
All present enzymatic activity of Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model):
7.1.2.2;
7.1.2.2;
Other elements in 8g0c:
The structure of Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model) also contains other interesting chemical elements:
| Magnesium | (Mg) | 4 atoms |
Bromine Binding Sites:
The binding sites of Bromine atom in the Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model)
(pdb code 8g0c). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 7 binding sites of Bromine where determined in the Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model), PDB code: 8g0c:
Jump to Bromine binding site number: 1; 2; 3; 4; 5; 6; 7;
In total 7 binding sites of Bromine where determined in the Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model), PDB code: 8g0c:
Jump to Bromine binding site number: 1; 2; 3; 4; 5; 6; 7;
Bromine binding site 1 out of 7 in 8g0c
Go back to
Bromine binding site 1 out
of 7 in the Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model)
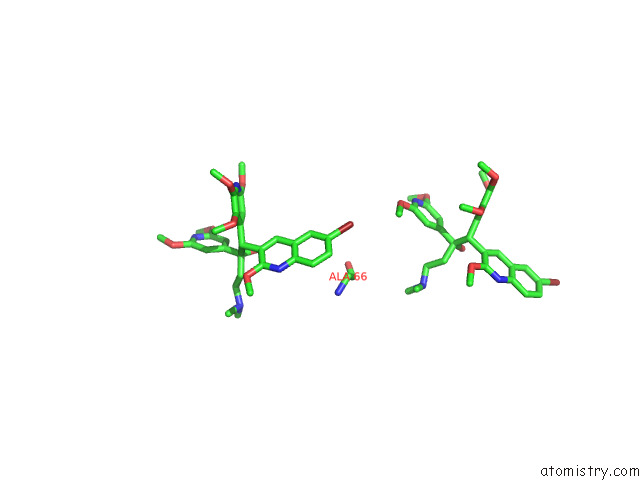
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model) within 5.0Å range:
|
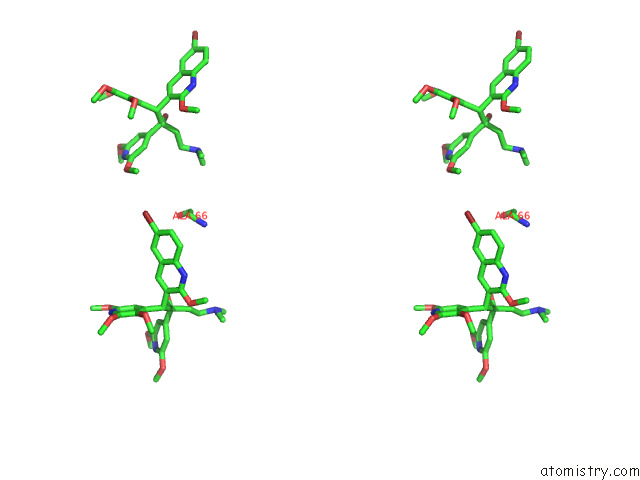
Bromine binding site 2 out of 7 in 8g0c
Go back to
Bromine binding site 2 out
of 7 in the Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model) within 5.0Å range:
|
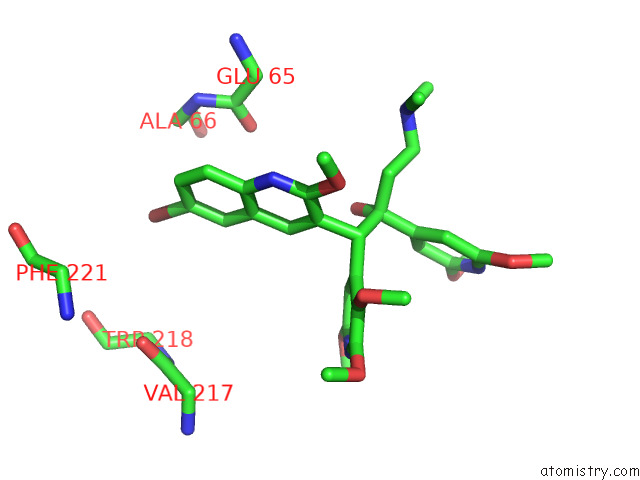
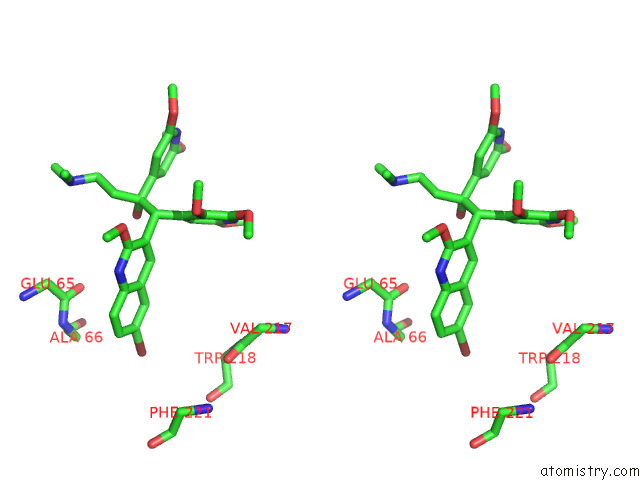
Bromine binding site 3 out of 7 in 8g0c
Go back to
Bromine binding site 3 out
of 7 in the Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model)
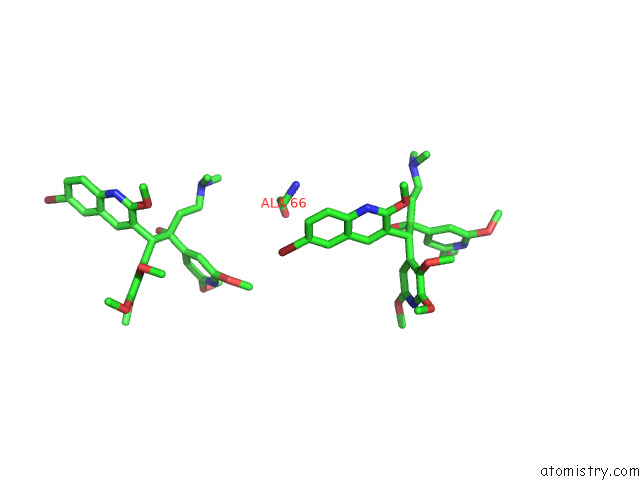
Mono view
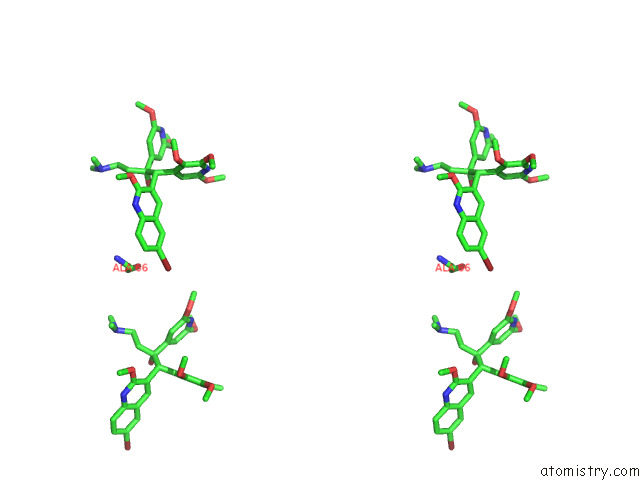
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model) within 5.0Å range:
|
Bromine binding site 4 out of 7 in 8g0c
Go back to
Bromine binding site 4 out
of 7 in the Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model)
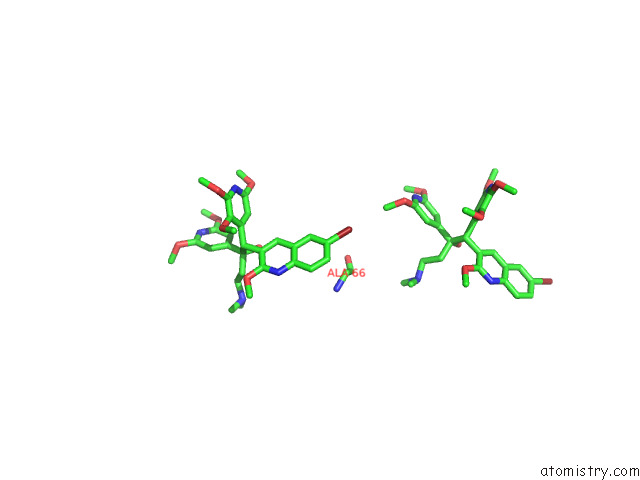
Mono view
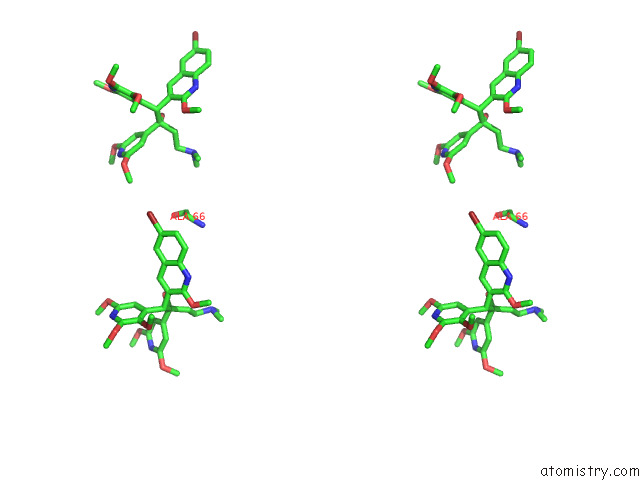
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 4 of Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model) within 5.0Å range:
|
Bromine binding site 5 out of 7 in 8g0c
Go back to
Bromine binding site 5 out
of 7 in the Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model)
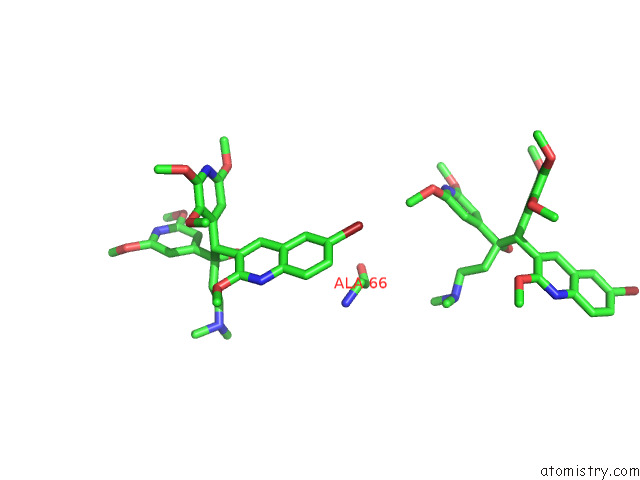
Mono view
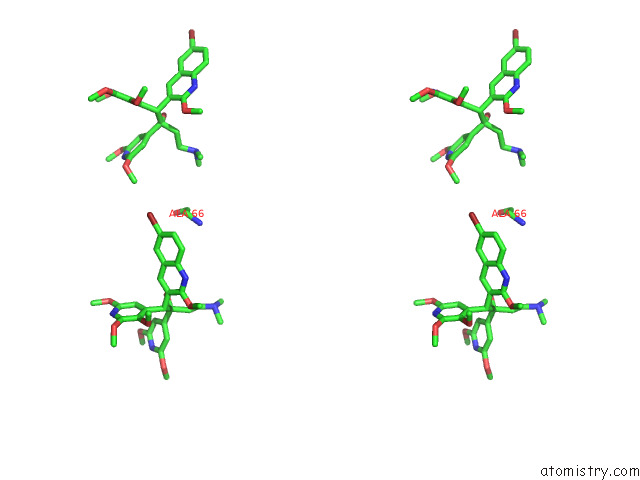
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 5 of Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model) within 5.0Å range:
|
Bromine binding site 6 out of 7 in 8g0c
Go back to
Bromine binding site 6 out
of 7 in the Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model)
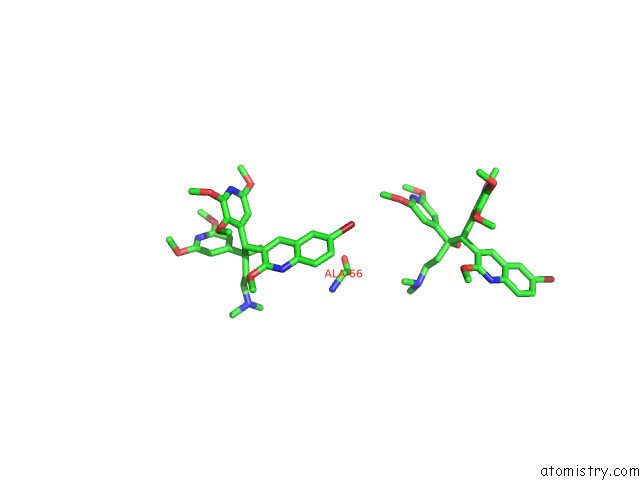
Mono view
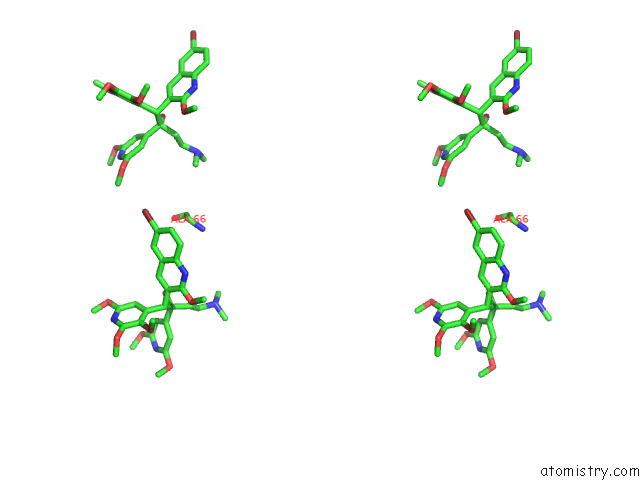
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 6 of Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model) within 5.0Å range:
|
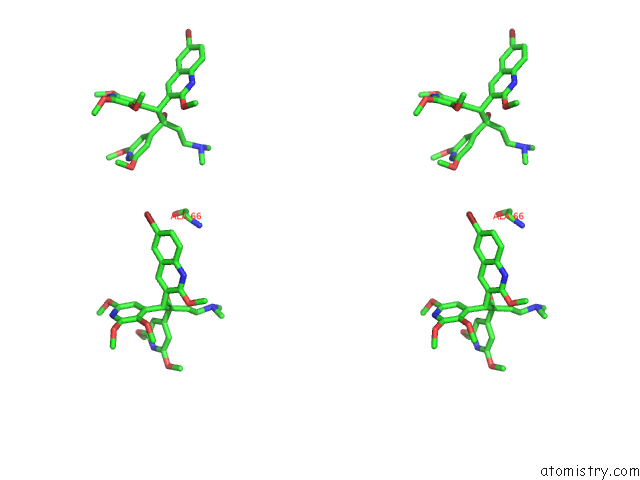
Bromine binding site 7 out of 7 in 8g0c
Go back to
Bromine binding site 7 out
of 7 in the Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model)
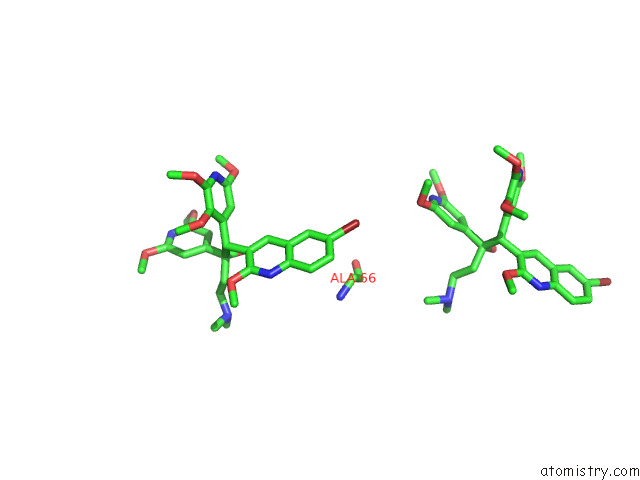
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 7 of Cryo-Em Structure of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model) within 5.0Å range:
|
Reference:
G.M.Courbon,
J.L.Rubinstein.
Structural Basis For Inhibition of Mycobacterial Atp Synthase By Squaramides and Second Generation Diarylquinolines To Be Published.
Page generated: Mon Jul 7 12:17:01 2025
Last articles
Cl in 3LATCl in 3L9R
Cl in 3LAK
Cl in 3L88
Cl in 3L9G
Cl in 3L7I
Cl in 3L7L
Cl in 3L7K
Cl in 3L6T
Cl in 3L6H