Bromine »
PDB 8gew-8k2l »
8hfk »
Bromine in PDB 8hfk: Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone
Protein crystallography data
The structure of Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone, PDB code: 8hfk
was solved by
X.D.Hou,
D.J.Yin,
Y.J.Rao,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.05 / 2.90 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 124.867, 124.867, 133.195, 90, 90, 90 |
| R / Rfree (%) | 20.2 / 28.9 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone
(pdb code 8hfk). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 8 binding sites of Bromine where determined in the Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone, PDB code: 8hfk:
Jump to Bromine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Bromine where determined in the Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone, PDB code: 8hfk:
Jump to Bromine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Bromine binding site 1 out of 8 in 8hfk
Go back to
Bromine binding site 1 out
of 8 in the Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone
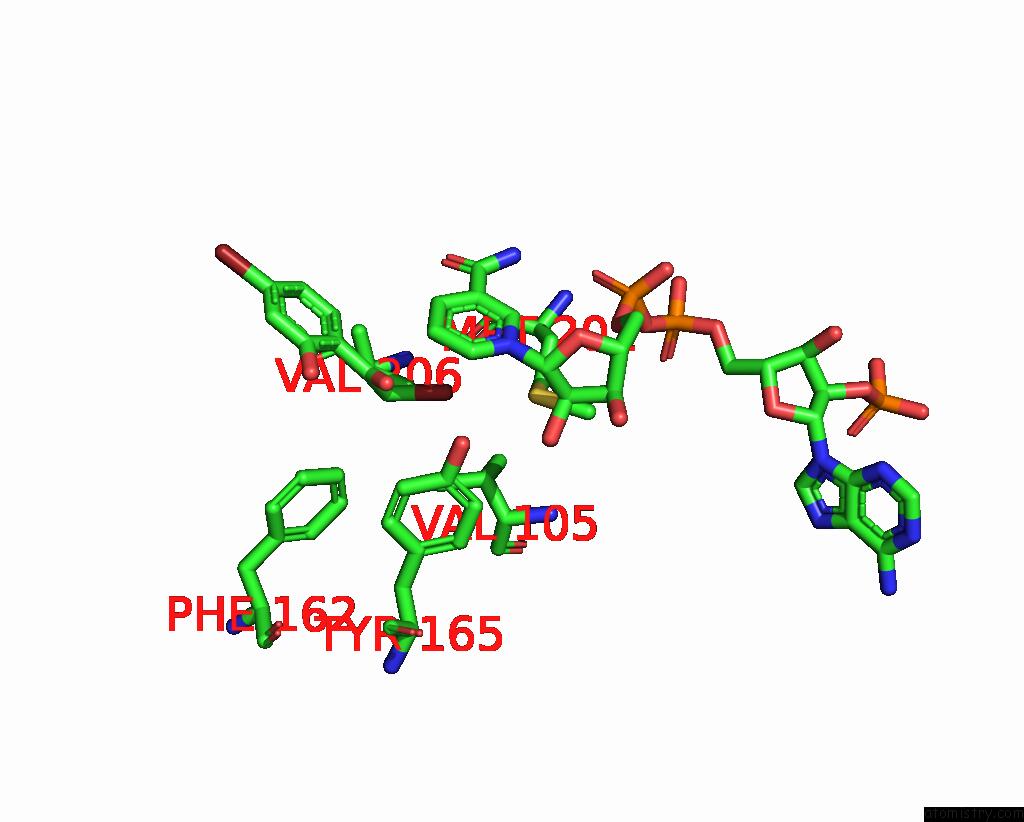
Mono view
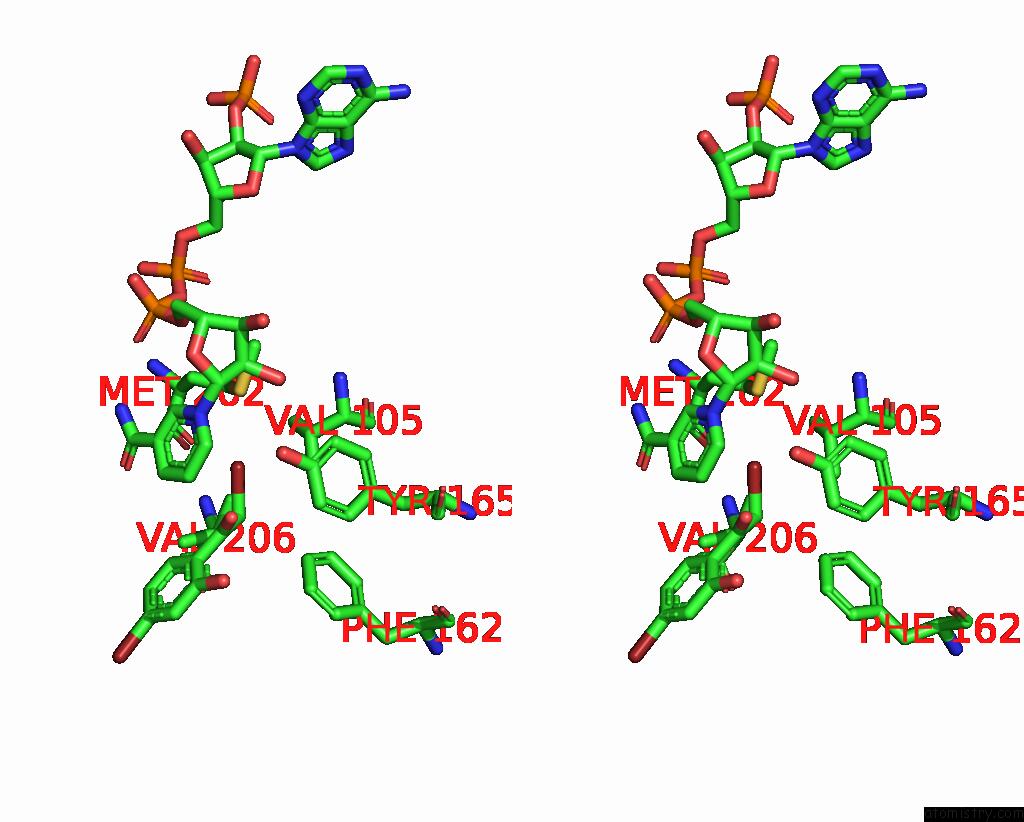
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone within 5.0Å range:
|
Bromine binding site 2 out of 8 in 8hfk
Go back to
Bromine binding site 2 out
of 8 in the Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone
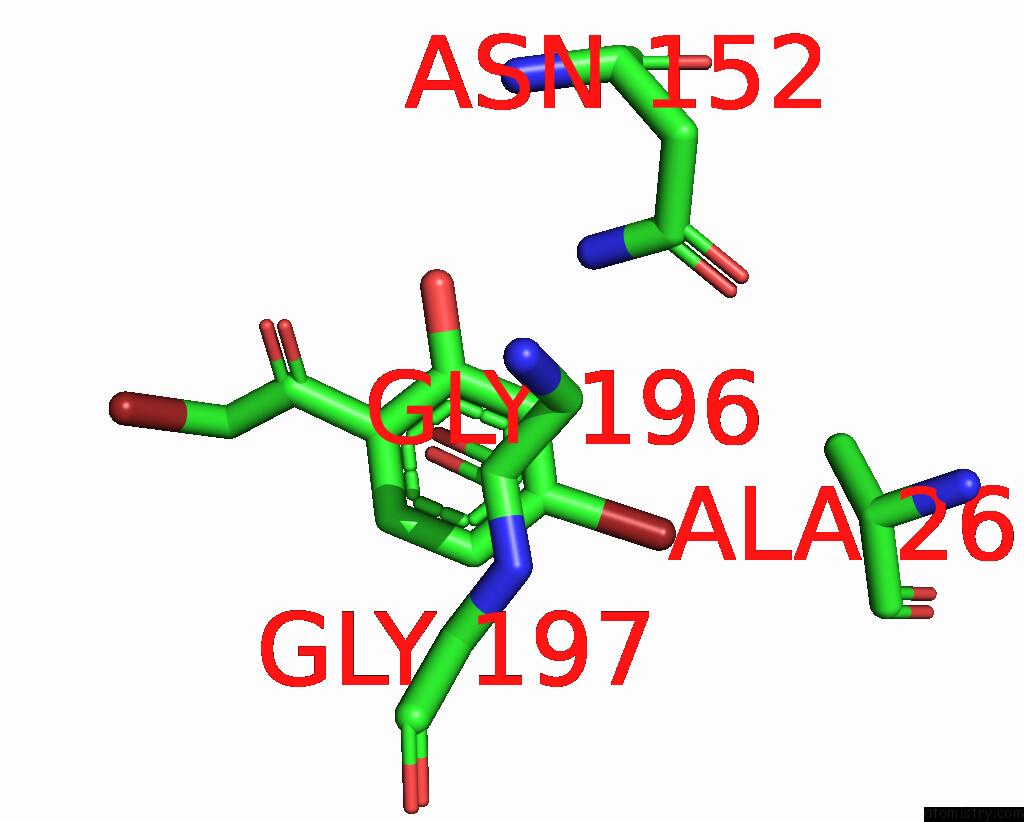
Mono view
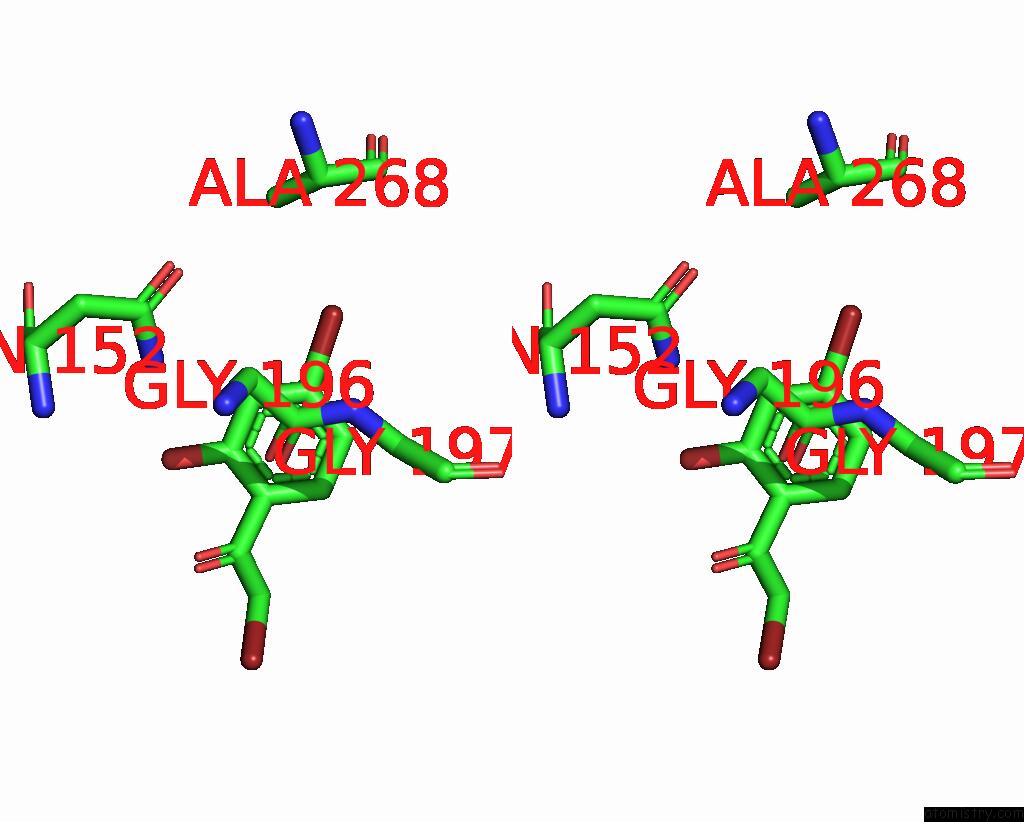
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone within 5.0Å range:
|
Bromine binding site 3 out of 8 in 8hfk
Go back to
Bromine binding site 3 out
of 8 in the Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone
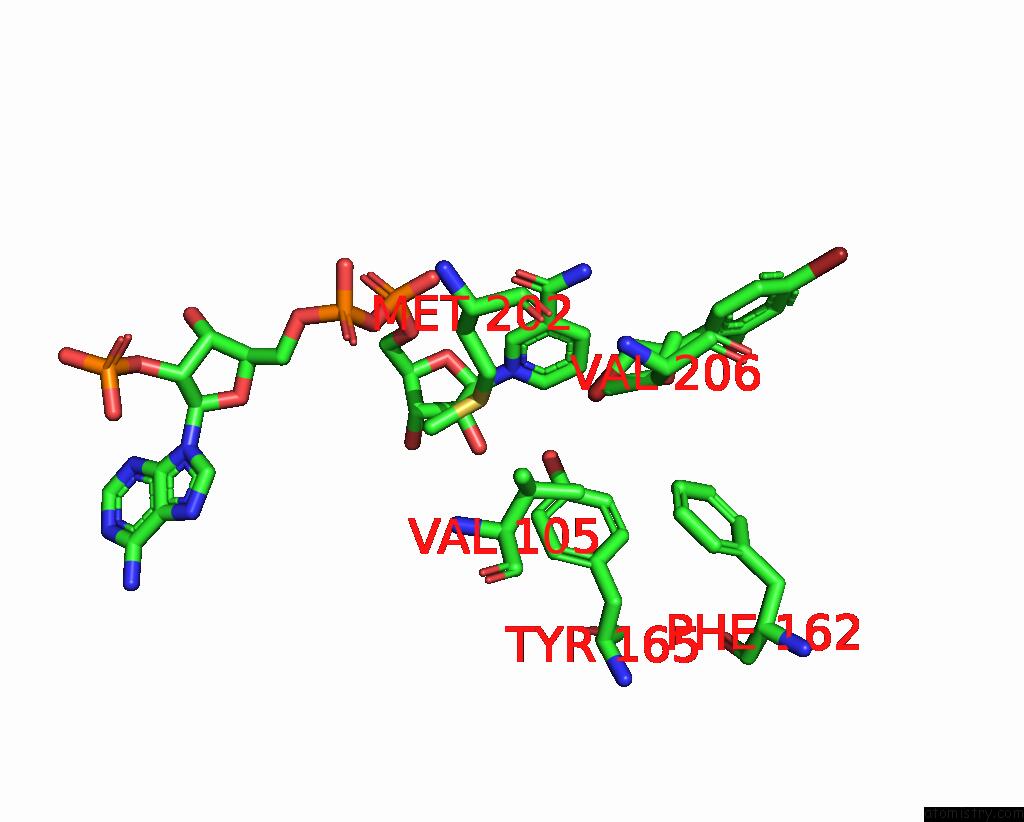
Mono view
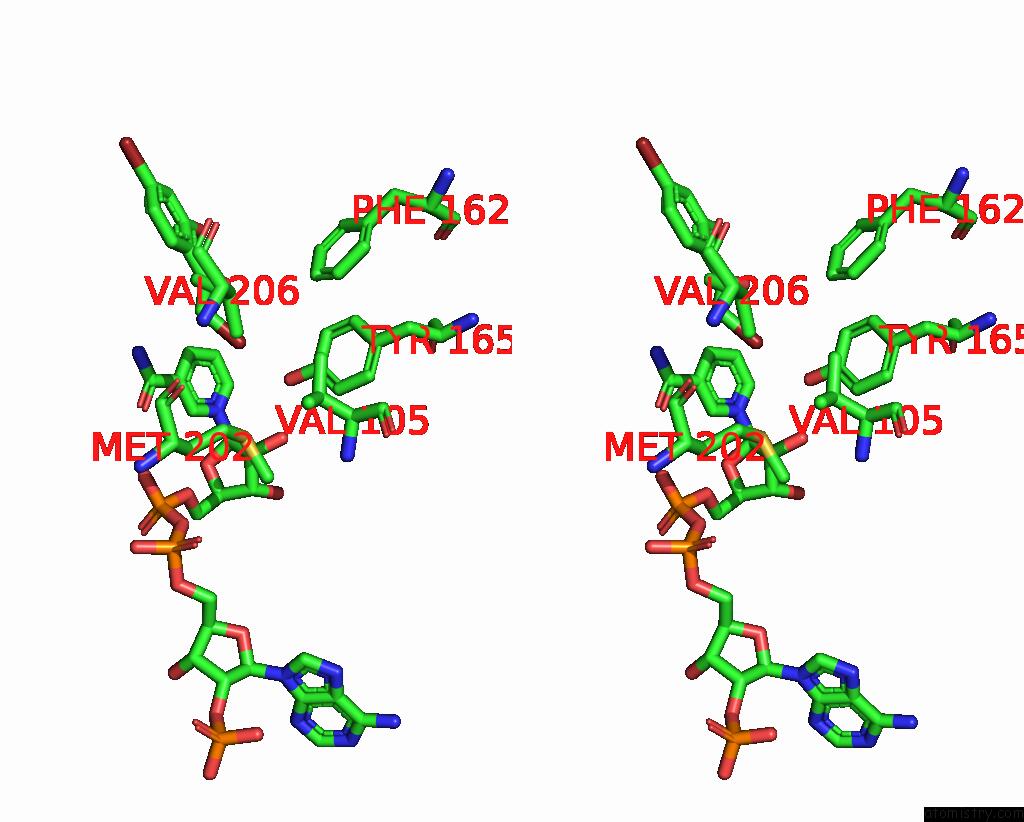
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone within 5.0Å range:
|
Bromine binding site 4 out of 8 in 8hfk
Go back to
Bromine binding site 4 out
of 8 in the Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone
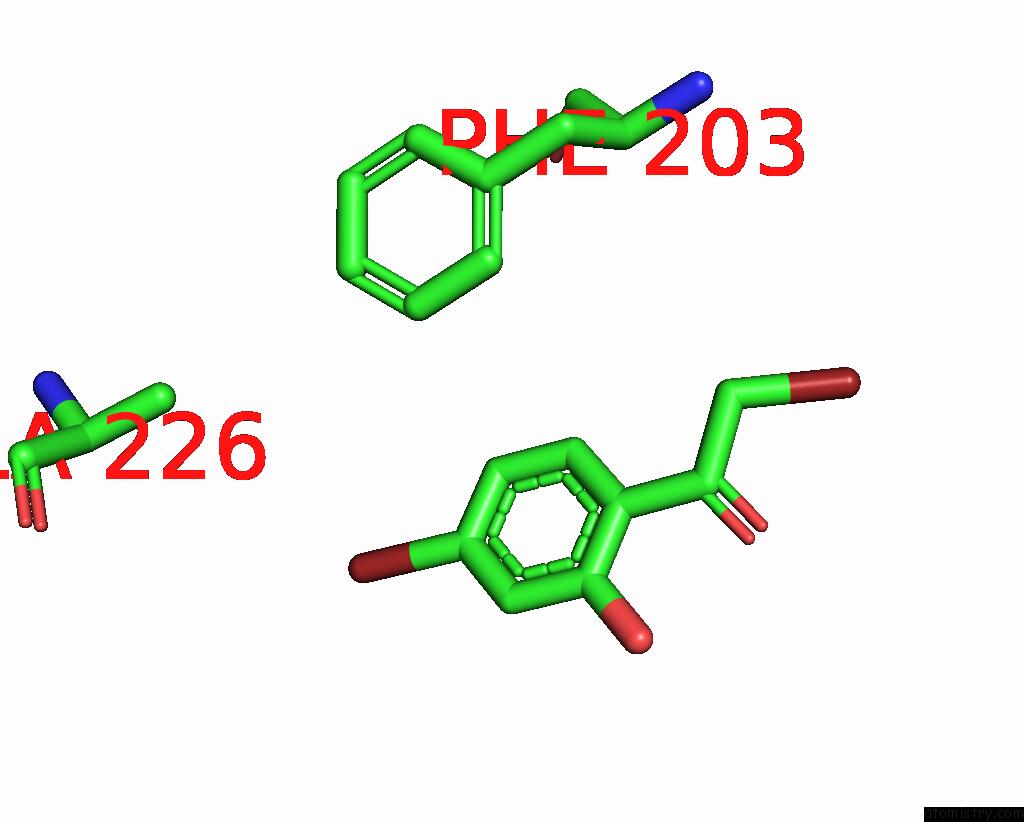
Mono view
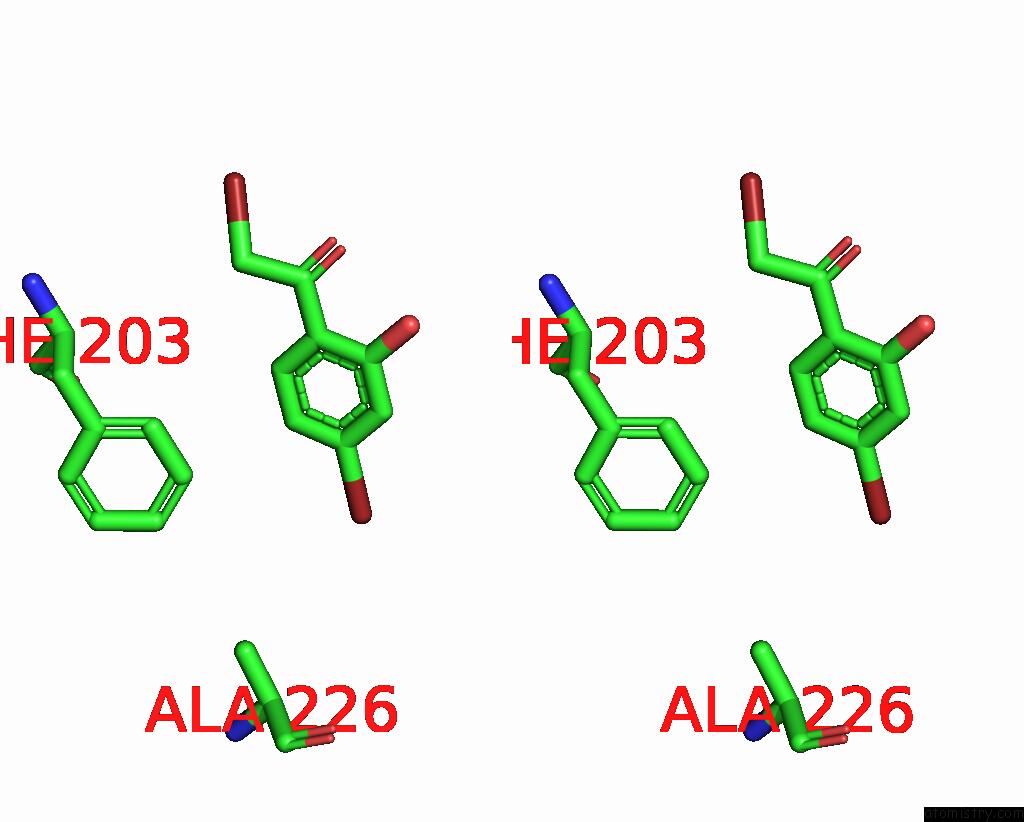
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 4 of Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone within 5.0Å range:
|
Bromine binding site 5 out of 8 in 8hfk
Go back to
Bromine binding site 5 out
of 8 in the Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone
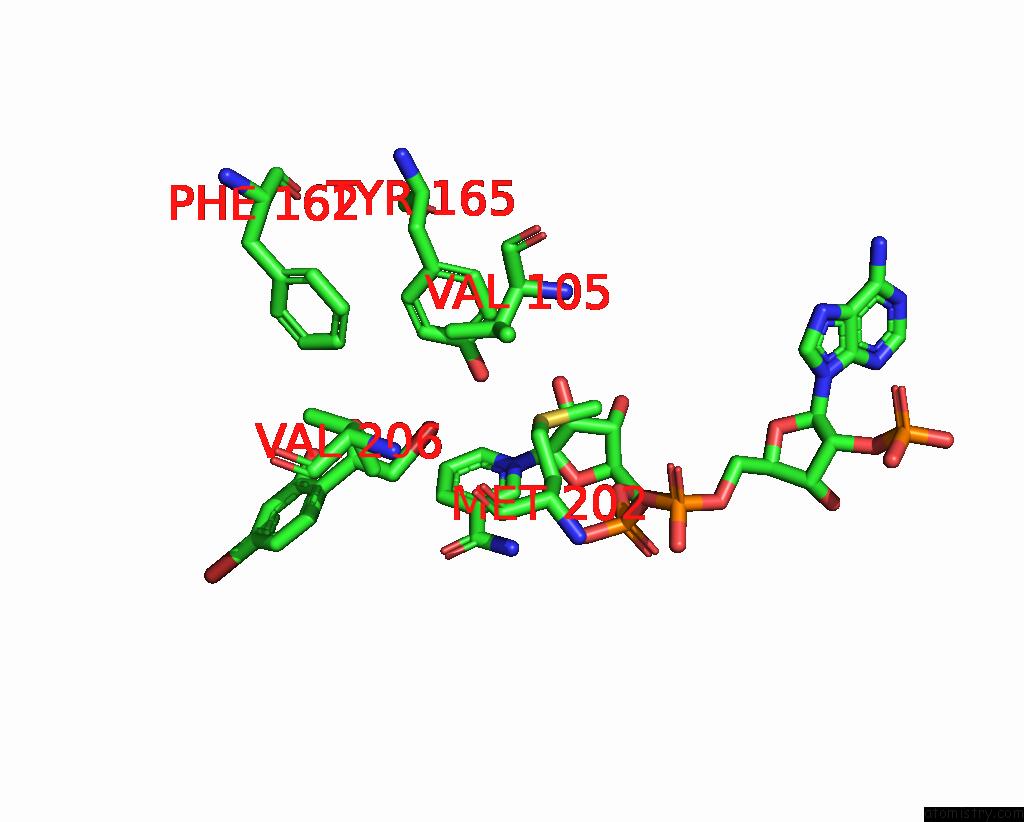
Mono view
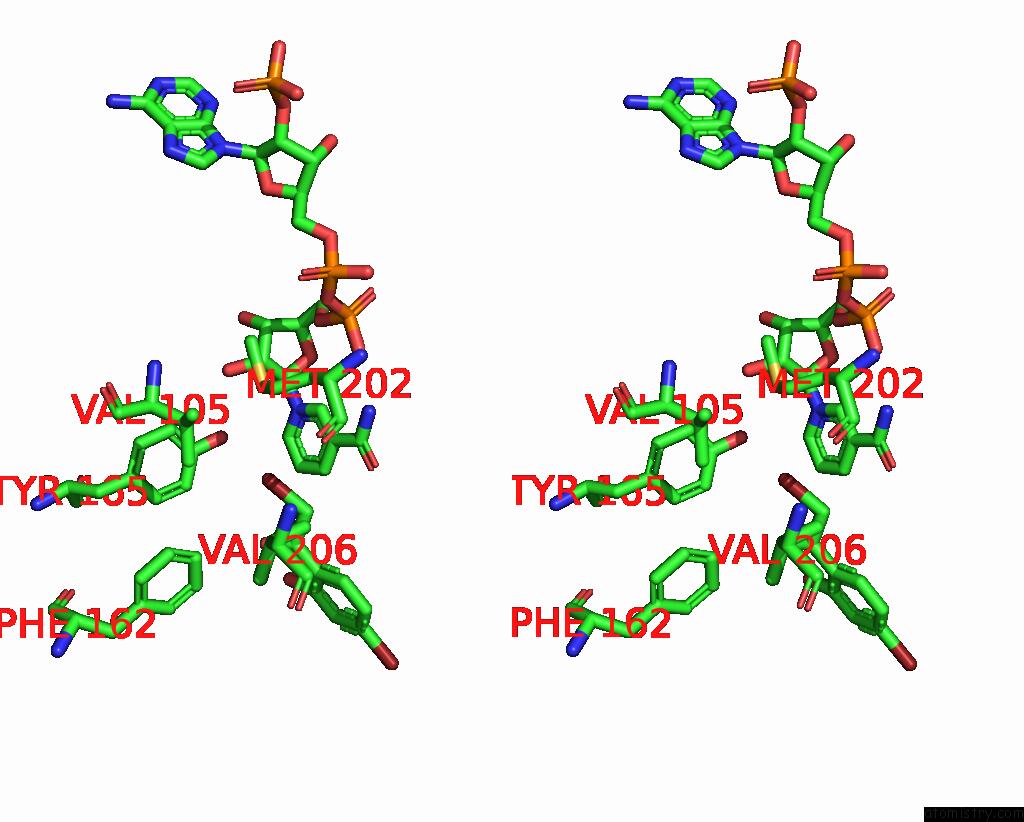
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 5 of Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone within 5.0Å range:
|
Bromine binding site 6 out of 8 in 8hfk
Go back to
Bromine binding site 6 out
of 8 in the Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone
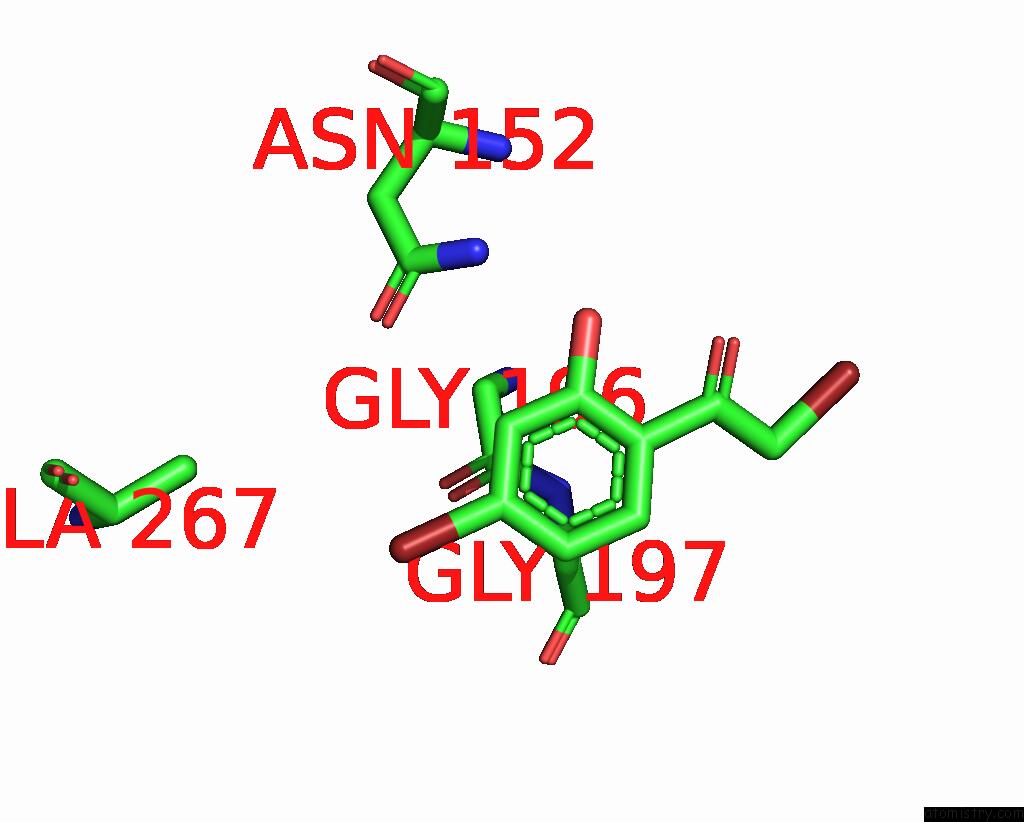
Mono view
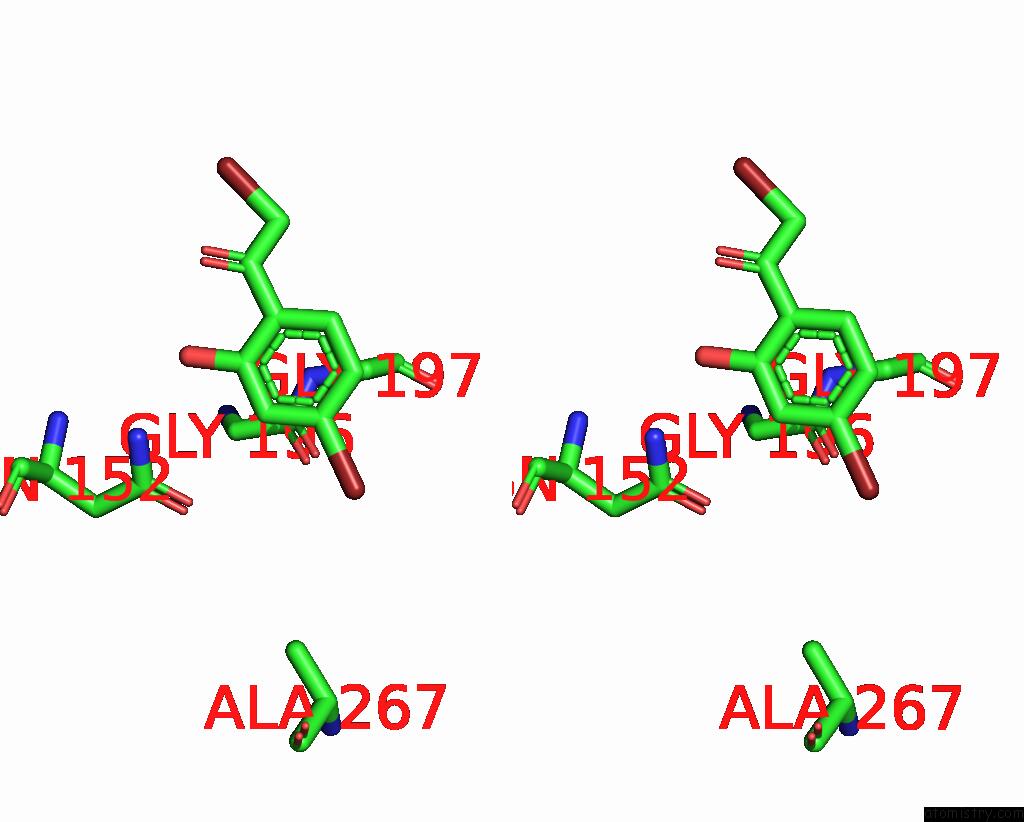
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 6 of Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone within 5.0Å range:
|
Bromine binding site 7 out of 8 in 8hfk
Go back to
Bromine binding site 7 out
of 8 in the Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone
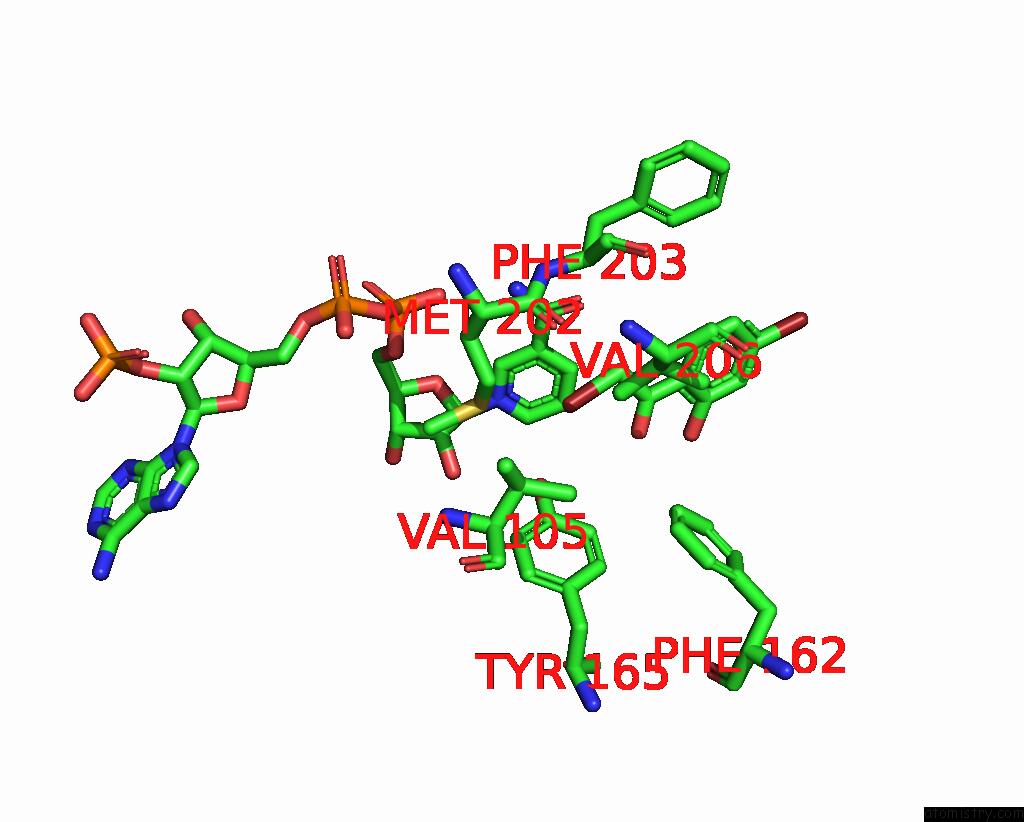
Mono view
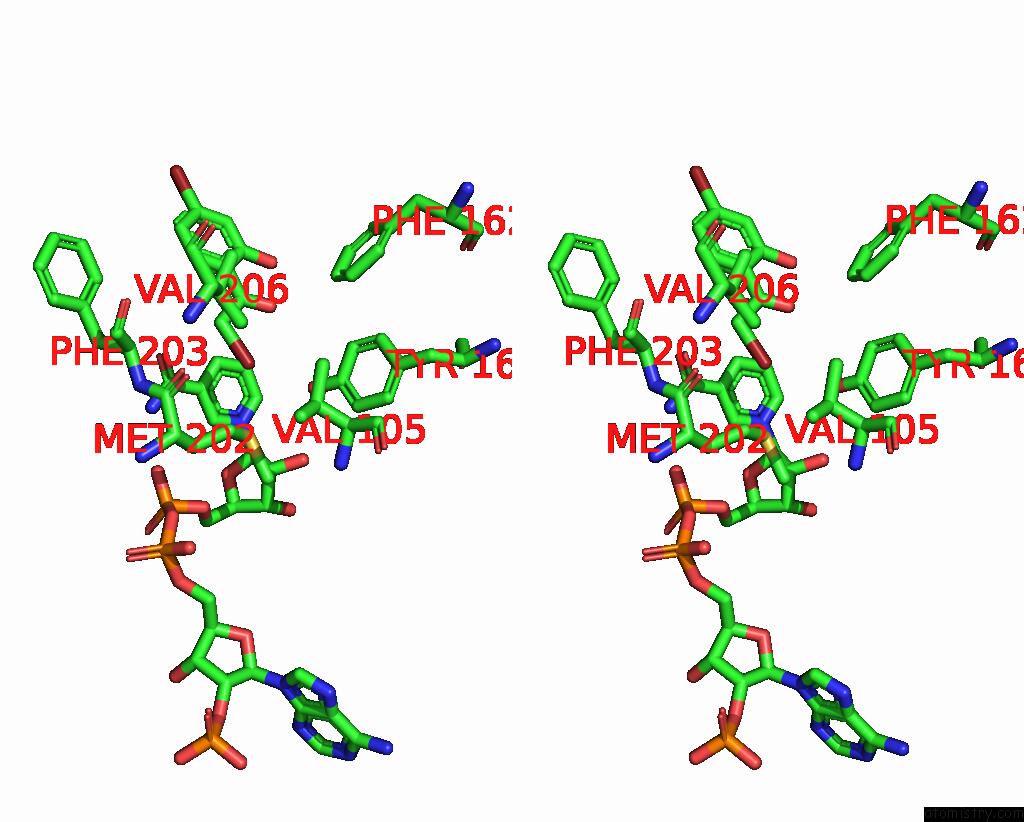
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 7 of Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone within 5.0Å range:
|
Bromine binding site 8 out of 8 in 8hfk
Go back to
Bromine binding site 8 out
of 8 in the Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone
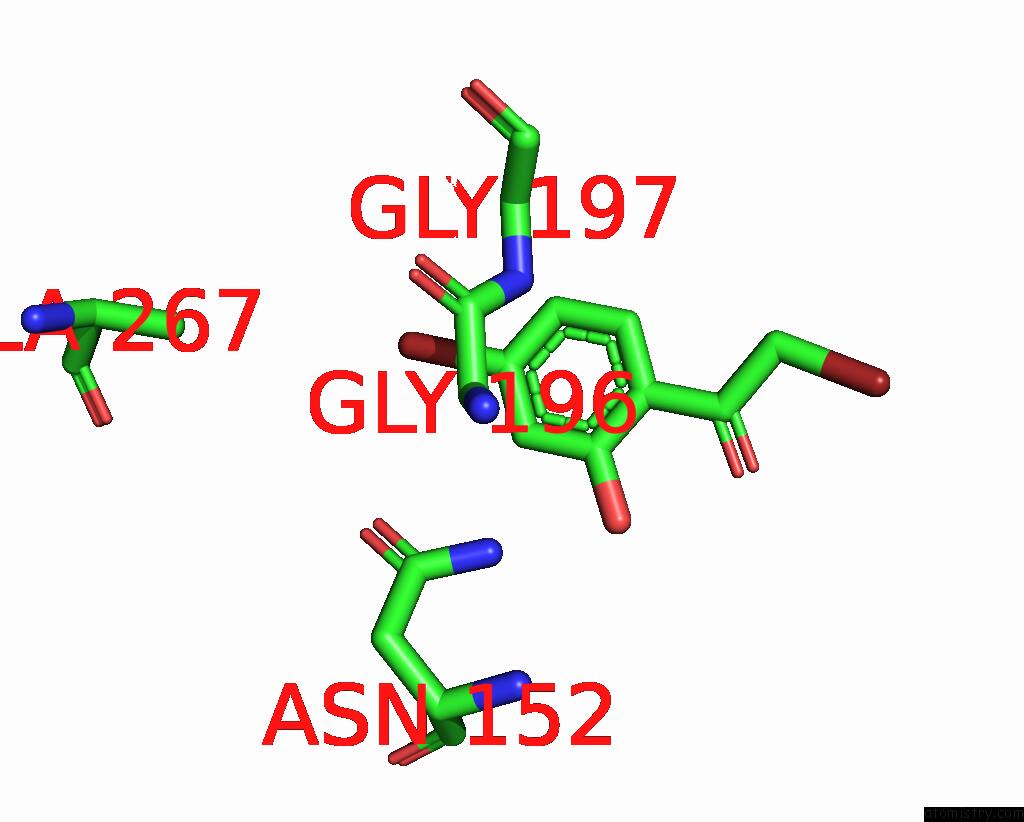
Mono view
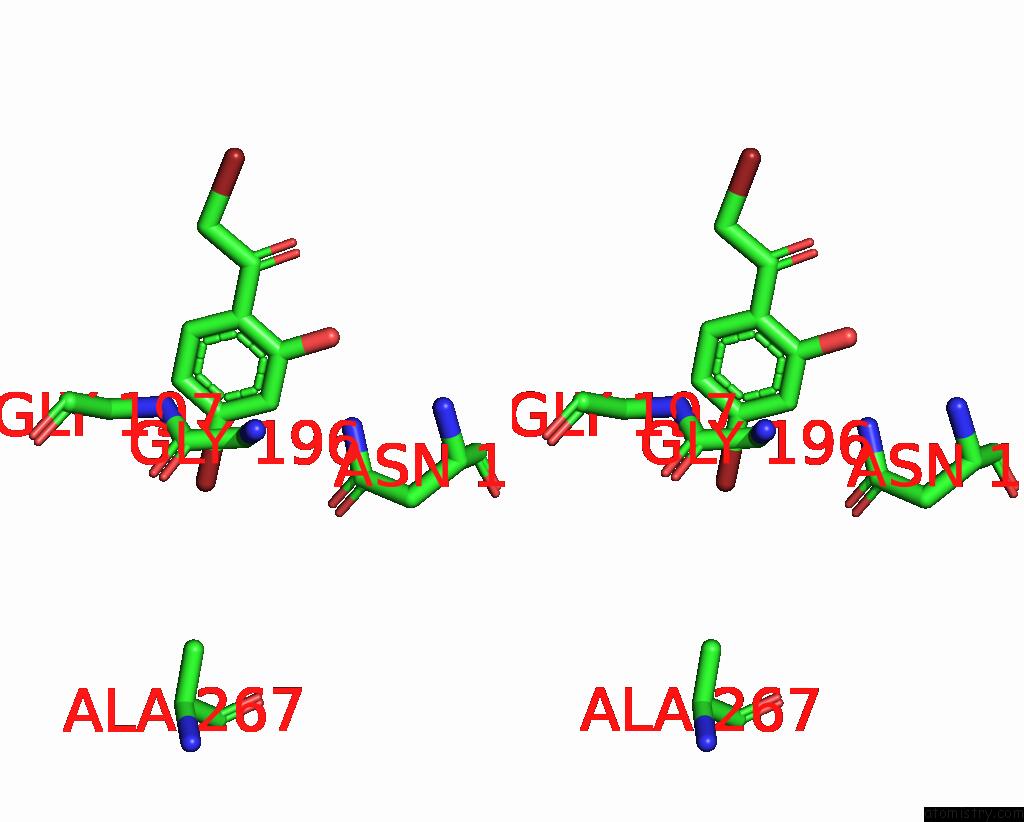
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 8 of Crystal Structure of Cbar Mutant (H162F) in Complex with Nadp+ and Halogenated Aryl Ketone within 5.0Å range:
|
Reference:
X.Hou,
H.Xu,
Z.Yuan,
Z.Deng,
K.Fu,
Y.Gao,
C.Liu,
Y.Zhang,
Y.Rao.
Structural Analysis of An Anthrol Reductase Inspires Enantioselective Synthesis of Enantiopure Hydroxycycloketones and Beta-Halohydrins. Nat Commun V. 14 353 2023.
ISSN: ESSN 2041-1723
PubMed: 36681664
DOI: 10.1038/S41467-023-36064-4
Page generated: Mon Jul 7 12:19:20 2025
ISSN: ESSN 2041-1723
PubMed: 36681664
DOI: 10.1038/S41467-023-36064-4
Last articles
Cl in 7Y11Cl in 7Y0B
Cl in 7XWR
Cl in 7XWQ
Cl in 7XWS
Cl in 7XVZ
Cl in 7XVM
Cl in 7XUL
Cl in 7XTQ
Cl in 7XUD