Bromine »
PDB 8k2m-8qs7 »
8oqt »
Bromine in PDB 8oqt: Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91
Enzymatic activity of Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91
All present enzymatic activity of Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91:
1.1.1.35;
1.1.1.35;
Protein crystallography data
The structure of Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91, PDB code: 8oqt
was solved by
S.Dalwani,
R.K.Wierenga,
R.Venkatesan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 54.59 / 2.62 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 247.476, 133.228, 115.928, 90, 109.65, 90 |
| R / Rfree (%) | 19.9 / 24.6 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91
(pdb code 8oqt). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 6 binding sites of Bromine where determined in the Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91, PDB code: 8oqt:
Jump to Bromine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Bromine where determined in the Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91, PDB code: 8oqt:
Jump to Bromine binding site number: 1; 2; 3; 4; 5; 6;
Bromine binding site 1 out of 6 in 8oqt
Go back to
Bromine binding site 1 out
of 6 in the Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91
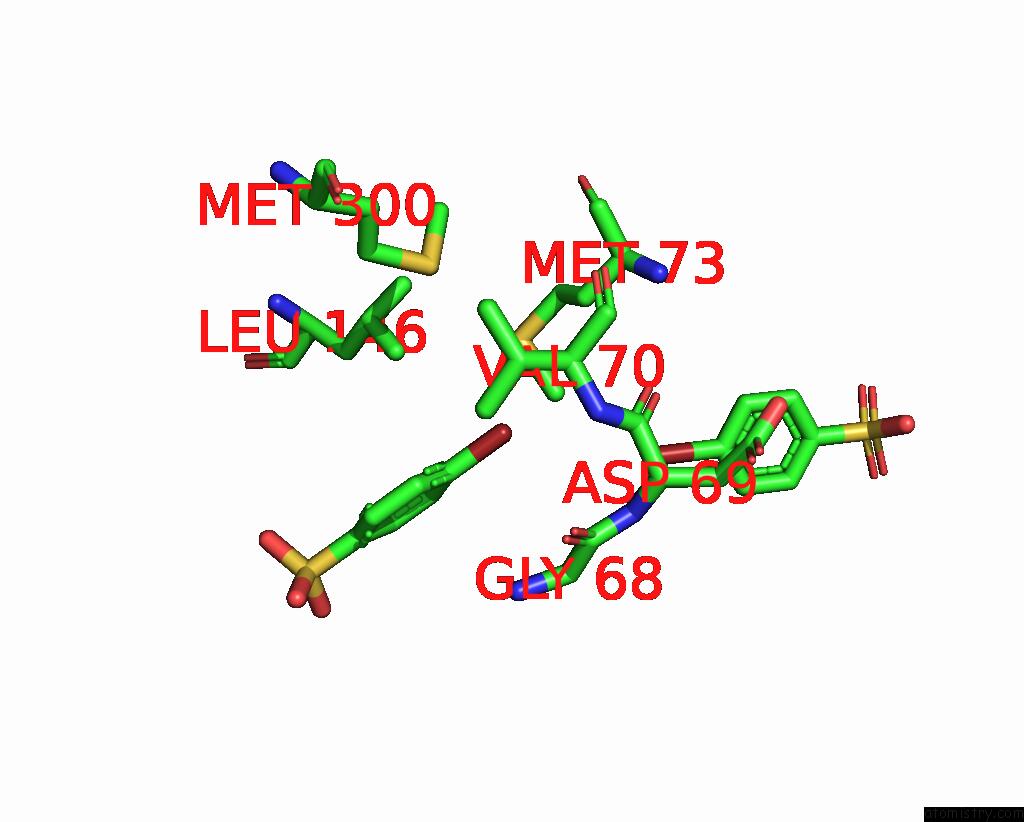
Mono view
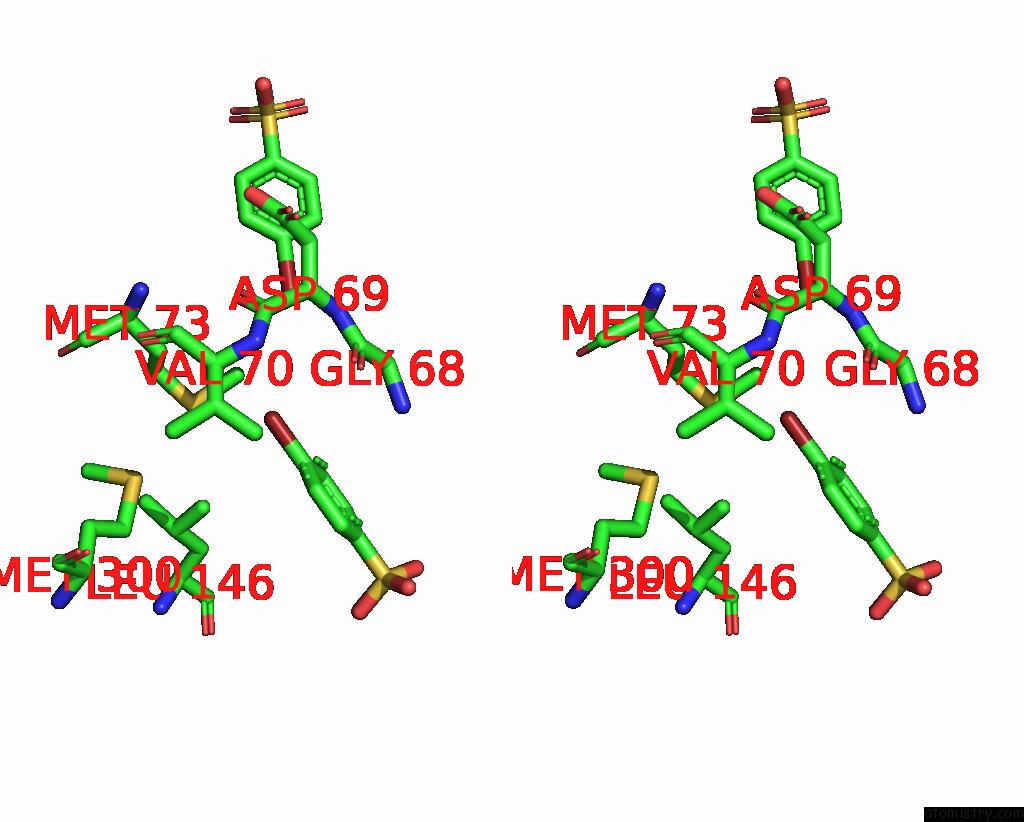
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91 within 5.0Å range:
|
Bromine binding site 2 out of 6 in 8oqt
Go back to
Bromine binding site 2 out
of 6 in the Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91
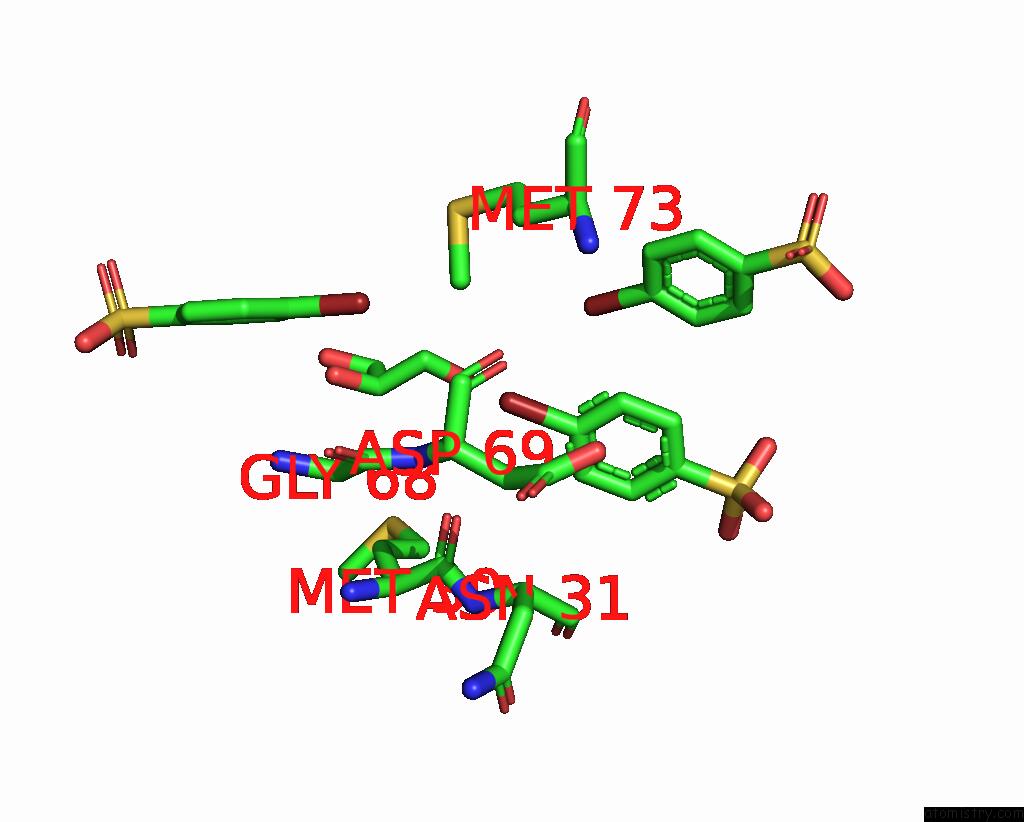
Mono view
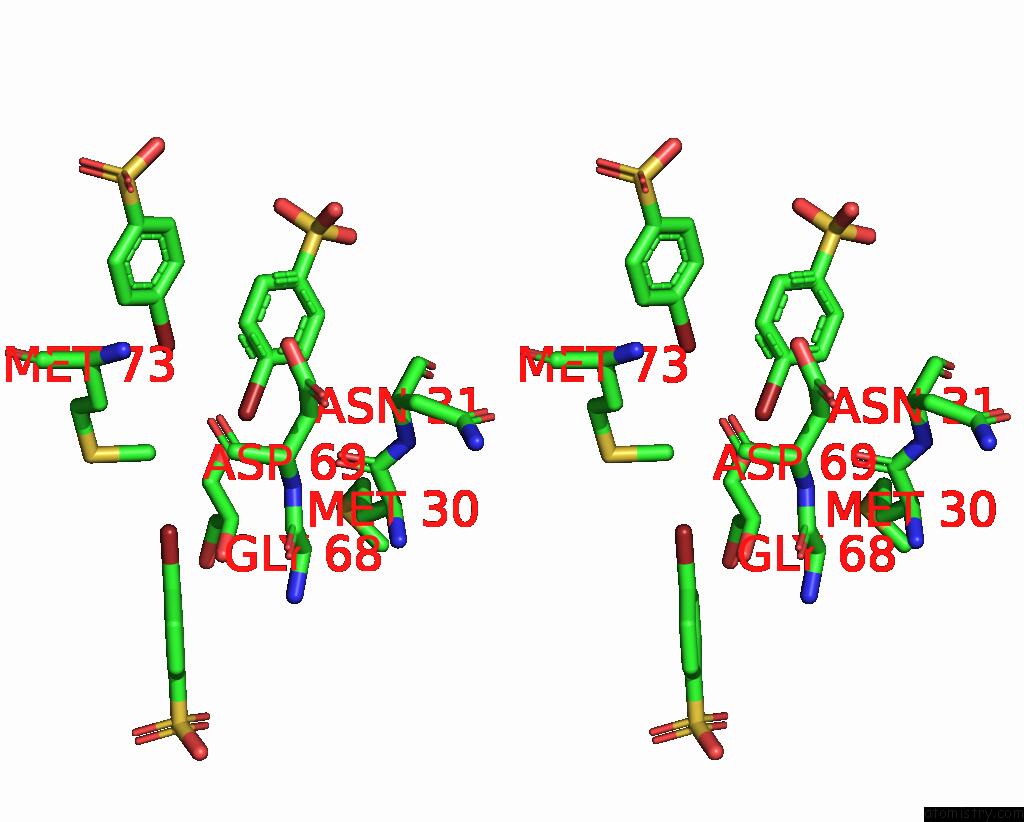
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91 within 5.0Å range:
|
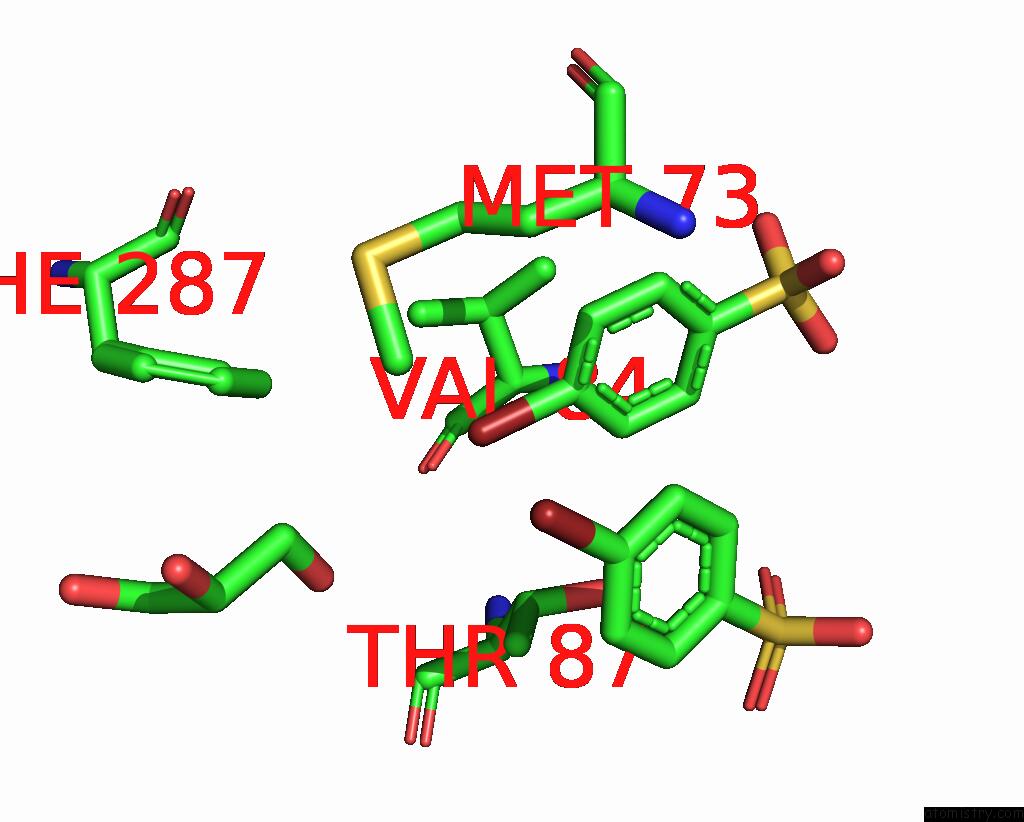
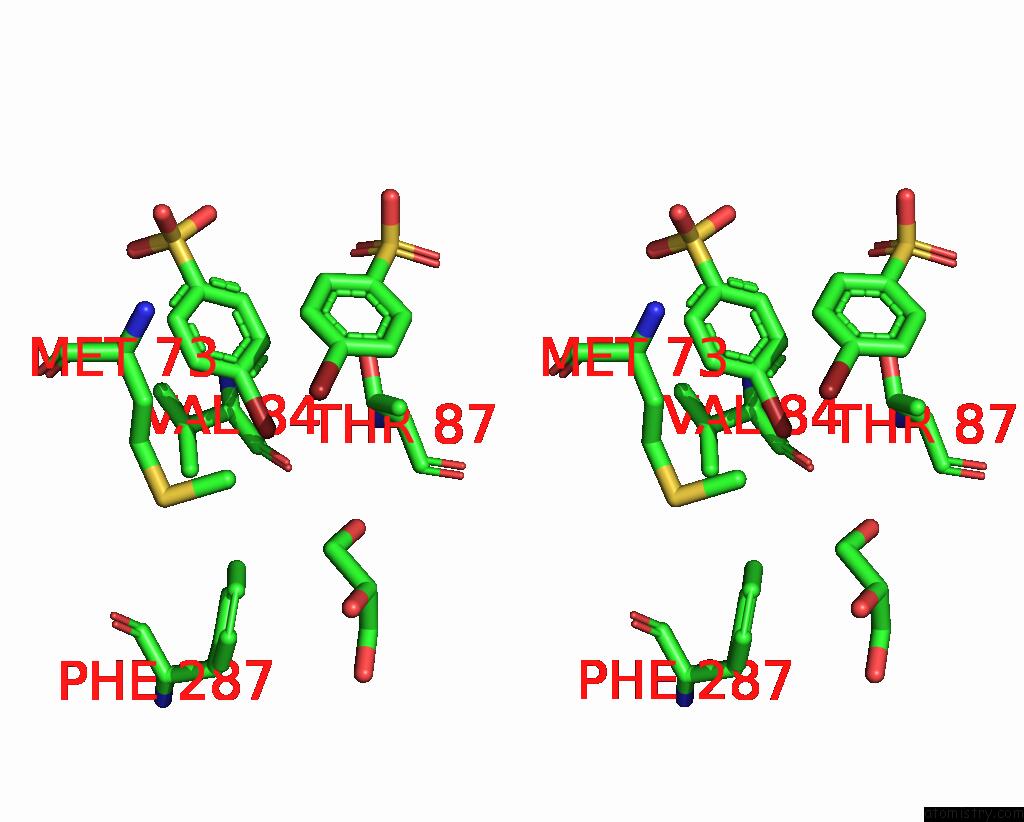
Bromine binding site 3 out of 6 in 8oqt
Go back to
Bromine binding site 3 out
of 6 in the Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91 within 5.0Å range:
|
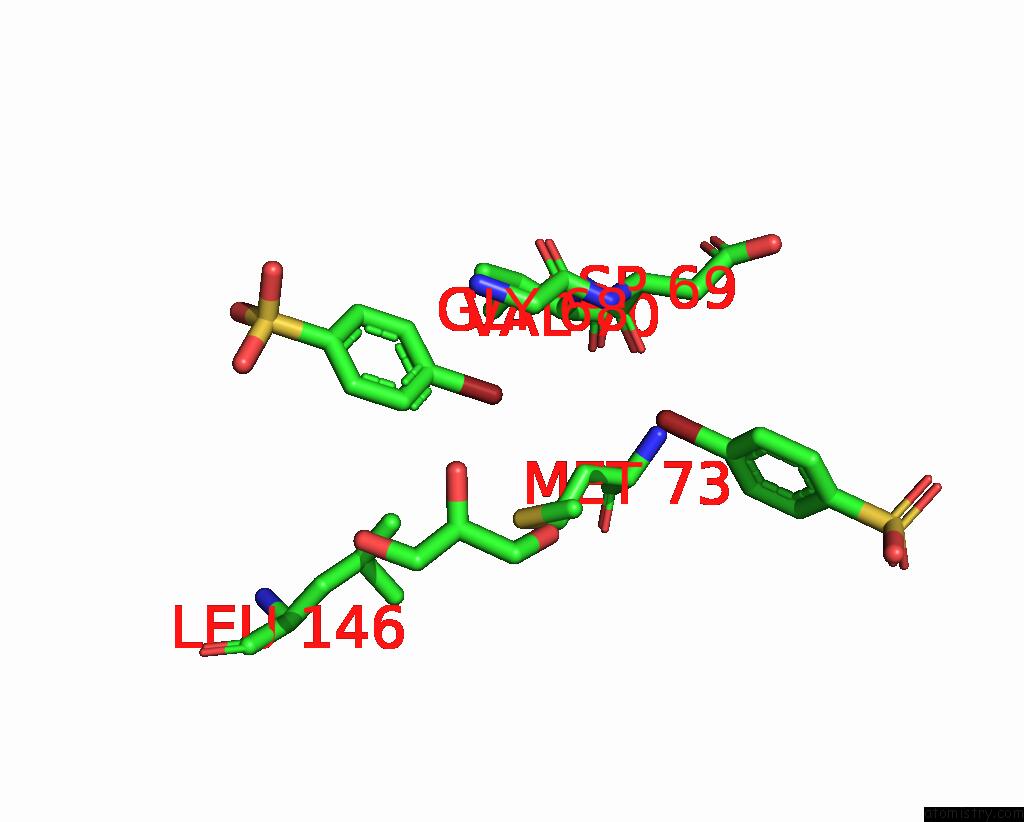
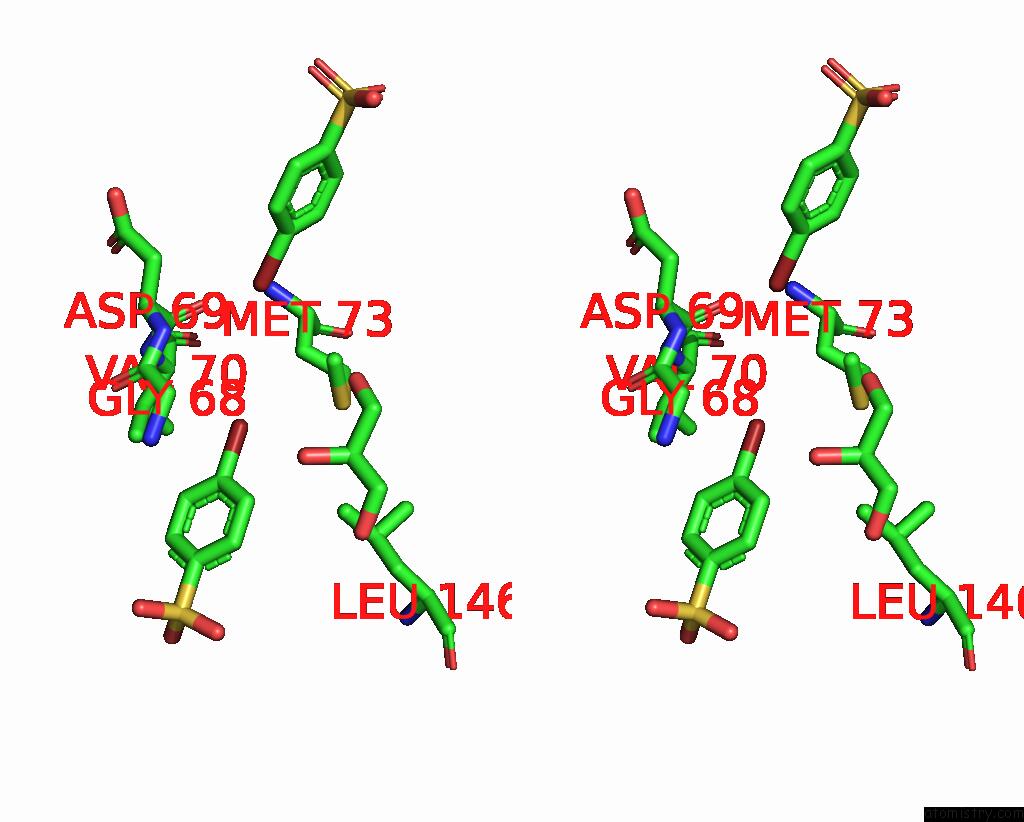
Bromine binding site 4 out of 6 in 8oqt
Go back to
Bromine binding site 4 out
of 6 in the Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 4 of Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91 within 5.0Å range:
|
Bromine binding site 5 out of 6 in 8oqt
Go back to
Bromine binding site 5 out
of 6 in the Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91
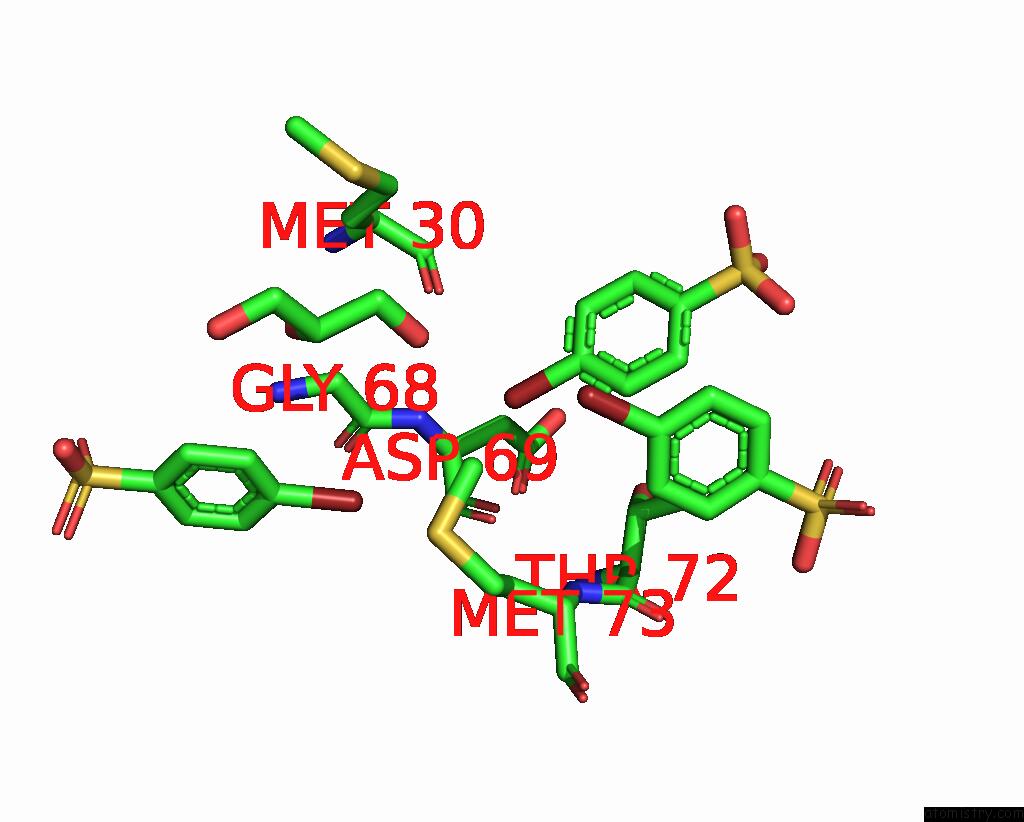
Mono view
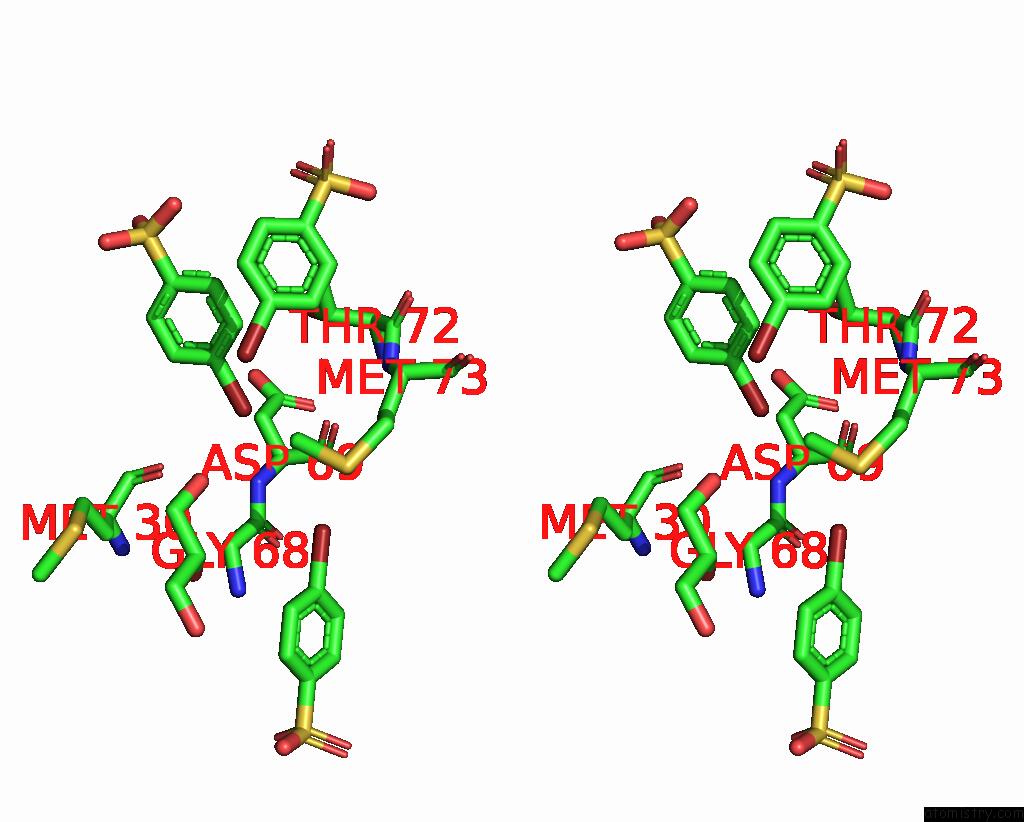
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 5 of Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91 within 5.0Å range:
|
Bromine binding site 6 out of 6 in 8oqt
Go back to
Bromine binding site 6 out
of 6 in the Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91
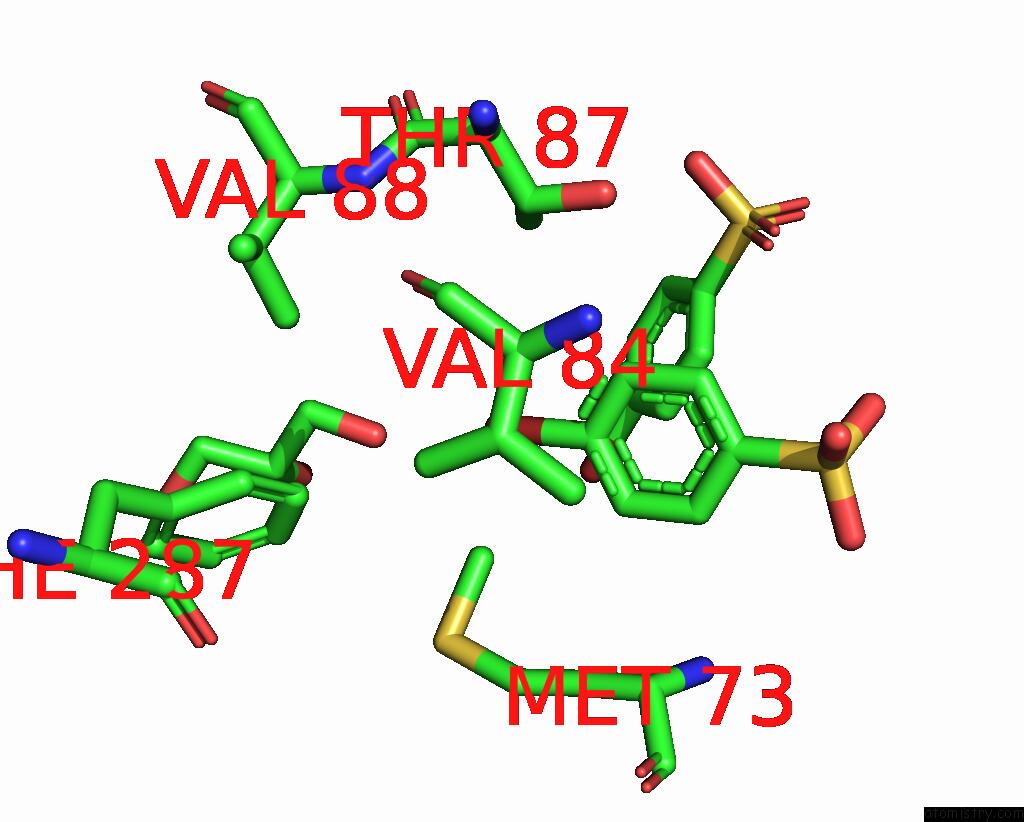
Mono view
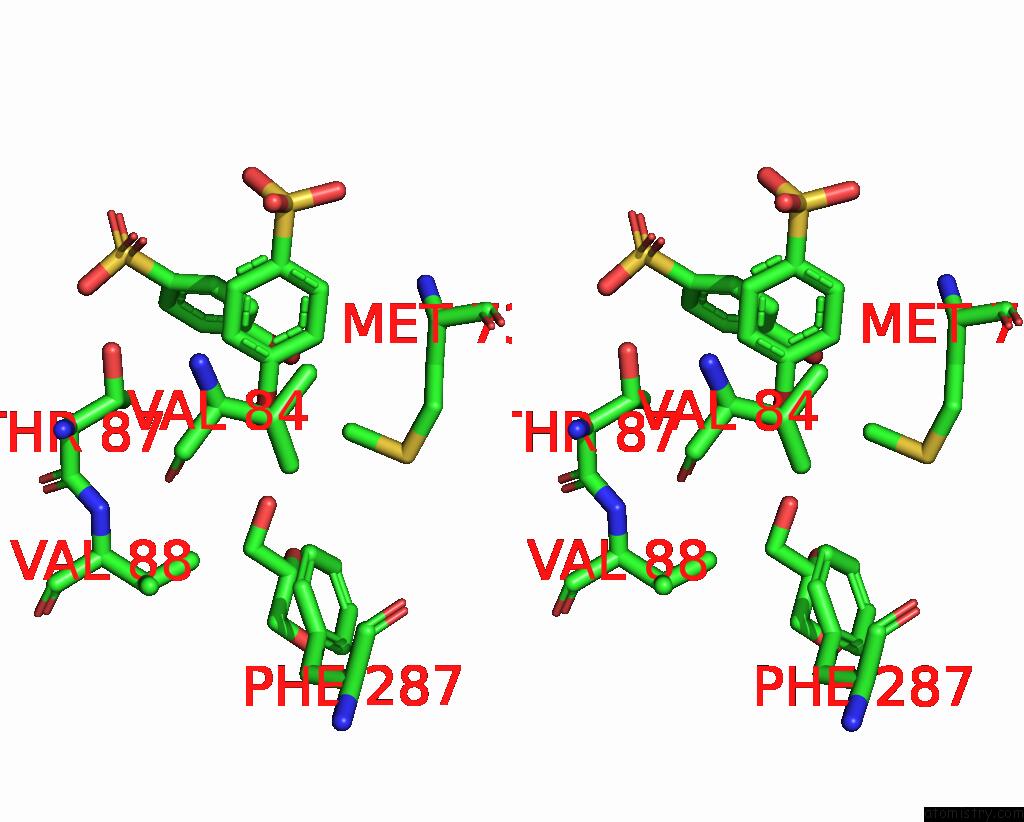
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 6 of Structure of Mycobacterium Tuberculosis Beta-Oxidation Trifunctional Enzyme in Complex with Fragment-M-91 within 5.0Å range:
|
Reference:
S.Dalwani,
A.Metz,
F.U.Huschmann,
M.S.Weiss,
R.K.Wierenga,
R.Venkatesan.
Crystallographic Fragment Binding Studies of the Mycobacterium Tuberculosis Trifunctional Enzyme Suggest Binding Pockets For the Tails of the Acyl-Coa Substrates at Its Active Sites and A Potential Substrate Channeling Path Between Them Biorxiv 2024.
ISSN: ISSN 2692-8205
DOI: 10.1101/2024.01.11.575214
Page generated: Mon Jul 7 12:28:35 2025
ISSN: ISSN 2692-8205
DOI: 10.1101/2024.01.11.575214
Last articles
Cl in 7ZD3Cl in 7ZD2
Cl in 7ZCZ
Cl in 7ZD1
Cl in 7ZD0
Cl in 7ZBV
Cl in 7ZCB
Cl in 7ZCY
Cl in 7ZBD
Cl in 7ZBB