Bromine »
PDB 8qs8-8u3a »
8r41 »
Bromine in PDB 8r41: Structure of CHI3L1 in Complex with Inhibitor 1
Protein crystallography data
The structure of Structure of CHI3L1 in Complex with Inhibitor 1, PDB code: 8r41
was solved by
E.Nowak,
A.Napiorkowska-Gromadzka,
M.Nowotny,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.57 / 2.25 |
| Space group | I 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 109.36, 122.25, 136.73, 90, 90, 90 |
| R / Rfree (%) | 20.5 / 23.4 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Structure of CHI3L1 in Complex with Inhibitor 1
(pdb code 8r41). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 4 binding sites of Bromine where determined in the Structure of CHI3L1 in Complex with Inhibitor 1, PDB code: 8r41:
Jump to Bromine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Bromine where determined in the Structure of CHI3L1 in Complex with Inhibitor 1, PDB code: 8r41:
Jump to Bromine binding site number: 1; 2; 3; 4;
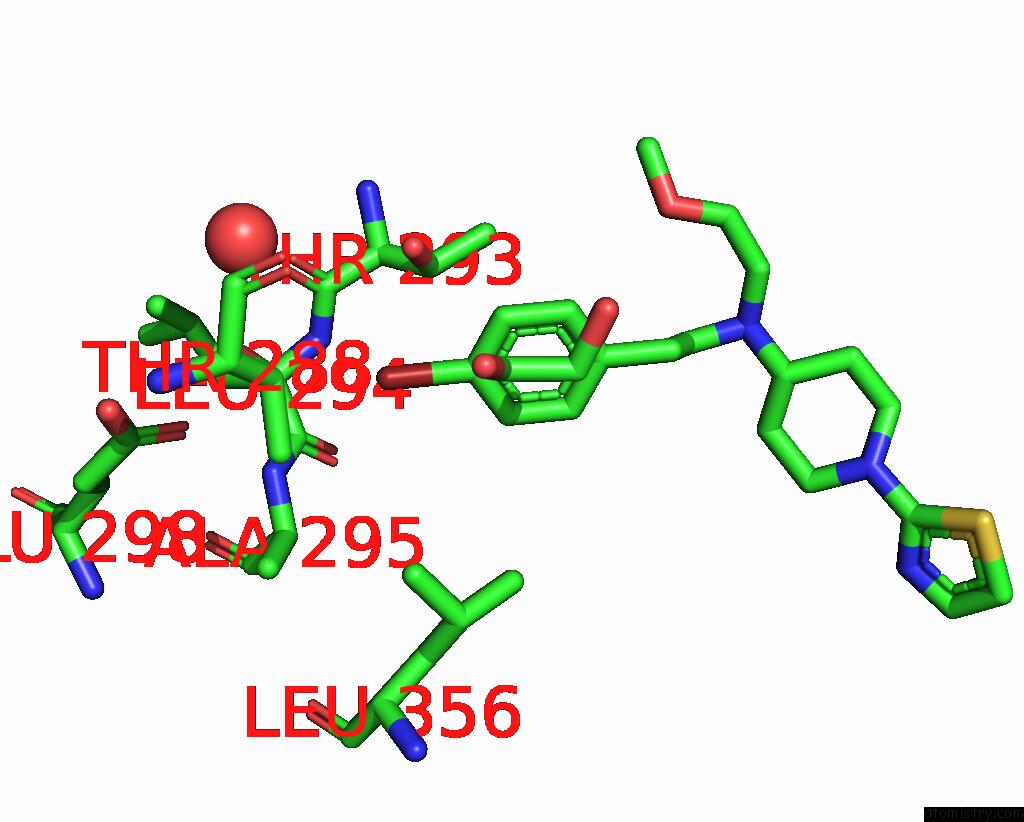
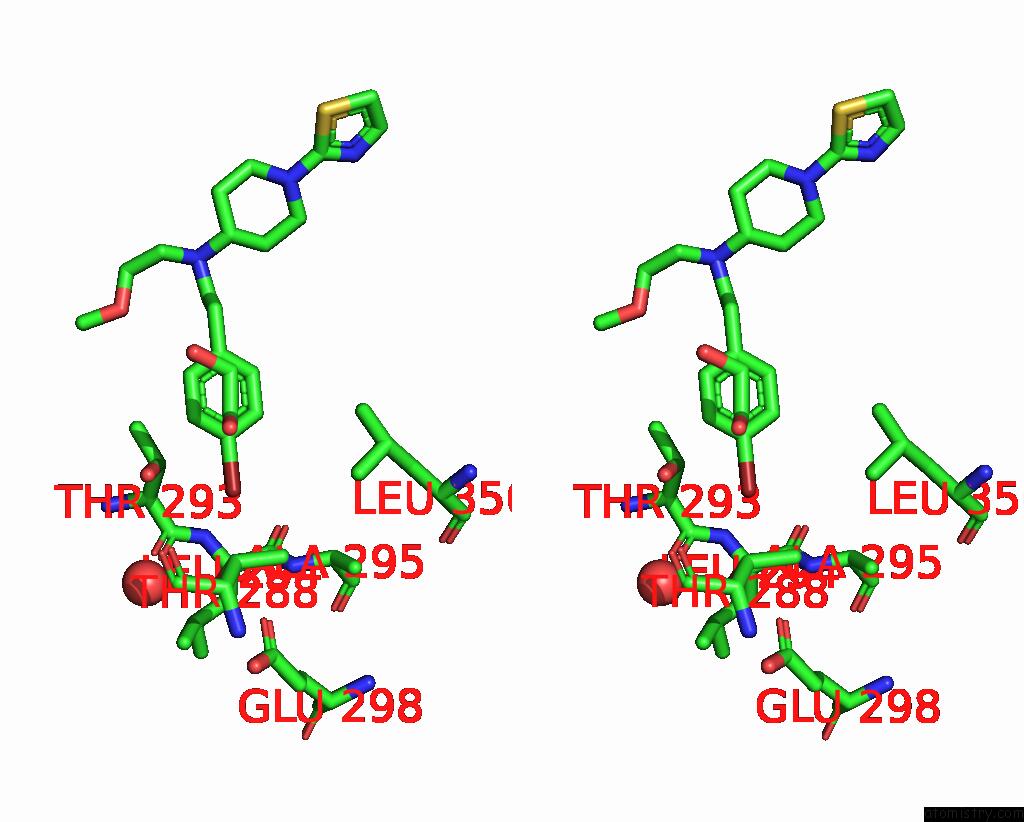
Bromine binding site 1 out of 4 in 8r41
Go back to
Bromine binding site 1 out
of 4 in the Structure of CHI3L1 in Complex with Inhibitor 1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Structure of CHI3L1 in Complex with Inhibitor 1 within 5.0Å range:
|
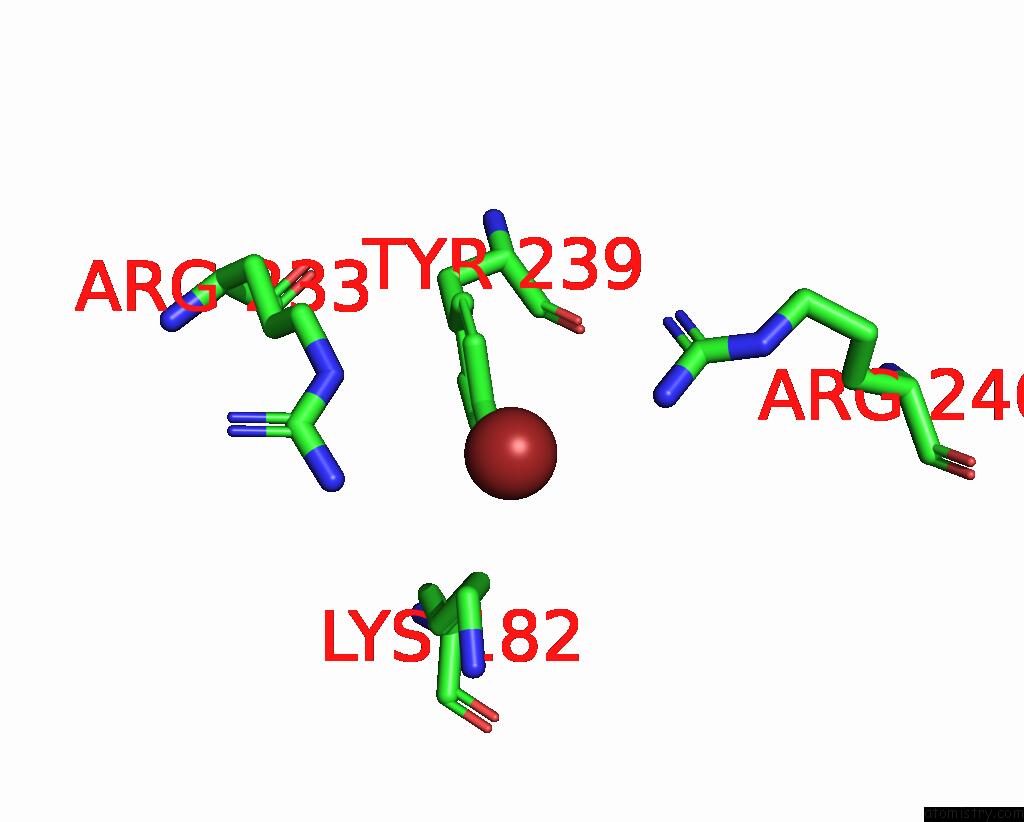
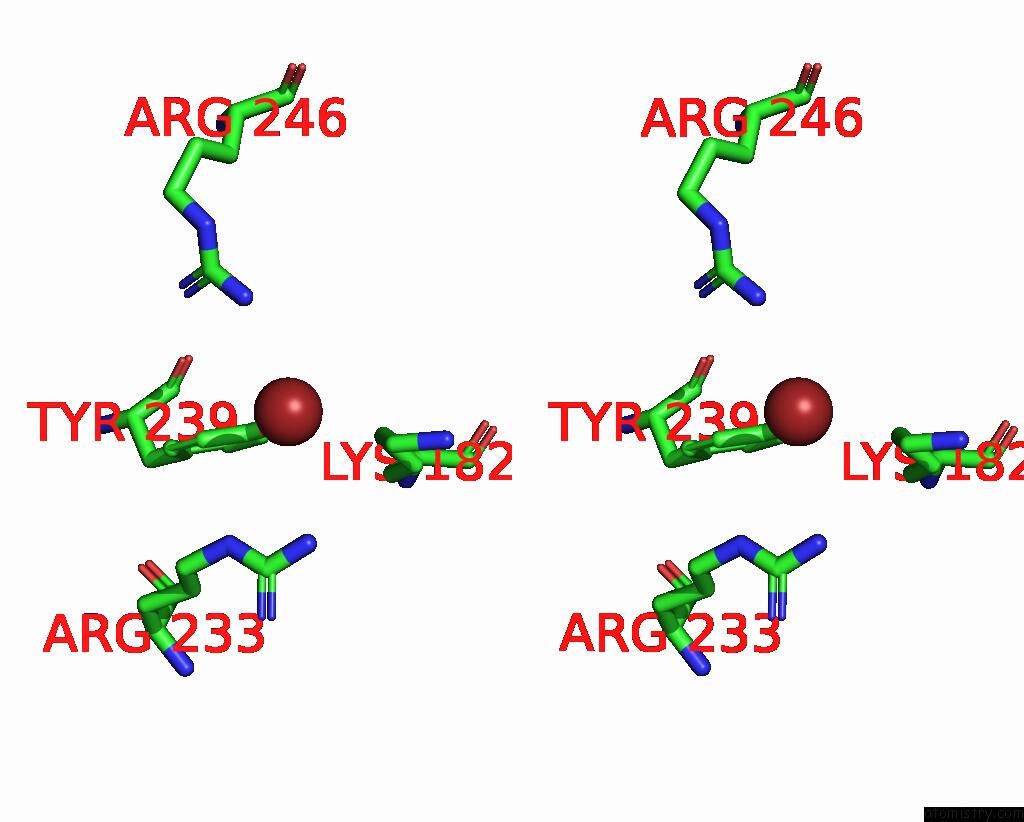
Bromine binding site 2 out of 4 in 8r41
Go back to
Bromine binding site 2 out
of 4 in the Structure of CHI3L1 in Complex with Inhibitor 1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Structure of CHI3L1 in Complex with Inhibitor 1 within 5.0Å range:
|
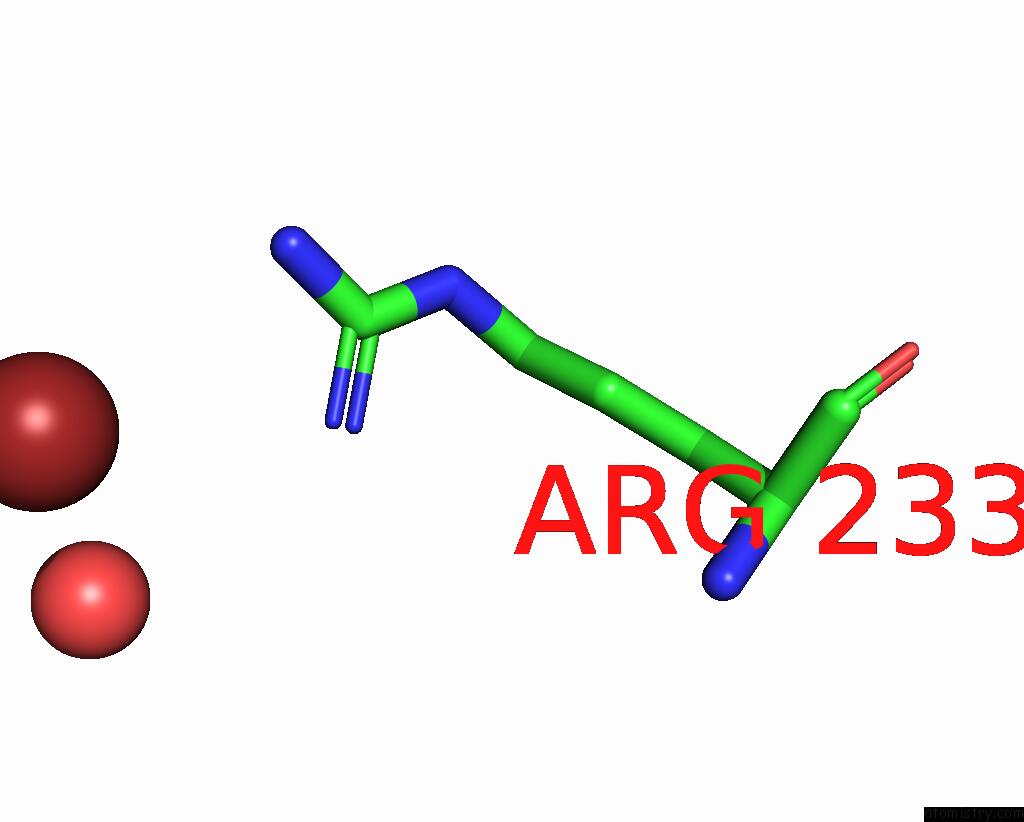
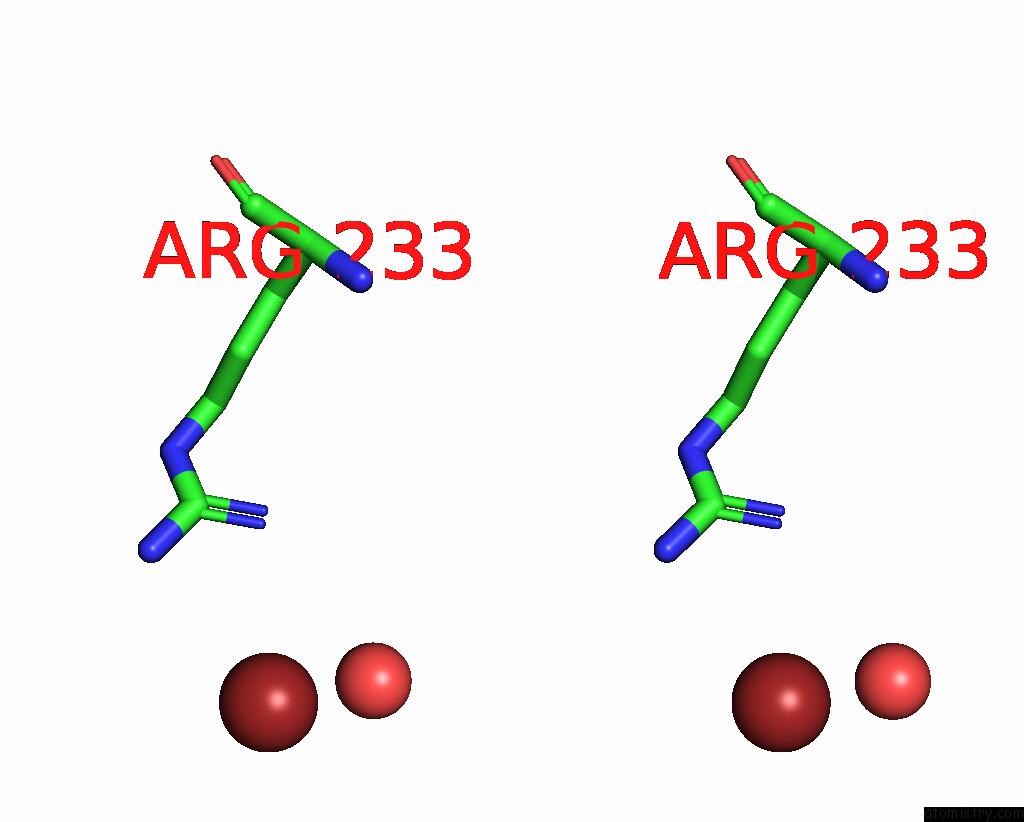
Bromine binding site 3 out of 4 in 8r41
Go back to
Bromine binding site 3 out
of 4 in the Structure of CHI3L1 in Complex with Inhibitor 1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of Structure of CHI3L1 in Complex with Inhibitor 1 within 5.0Å range:
|
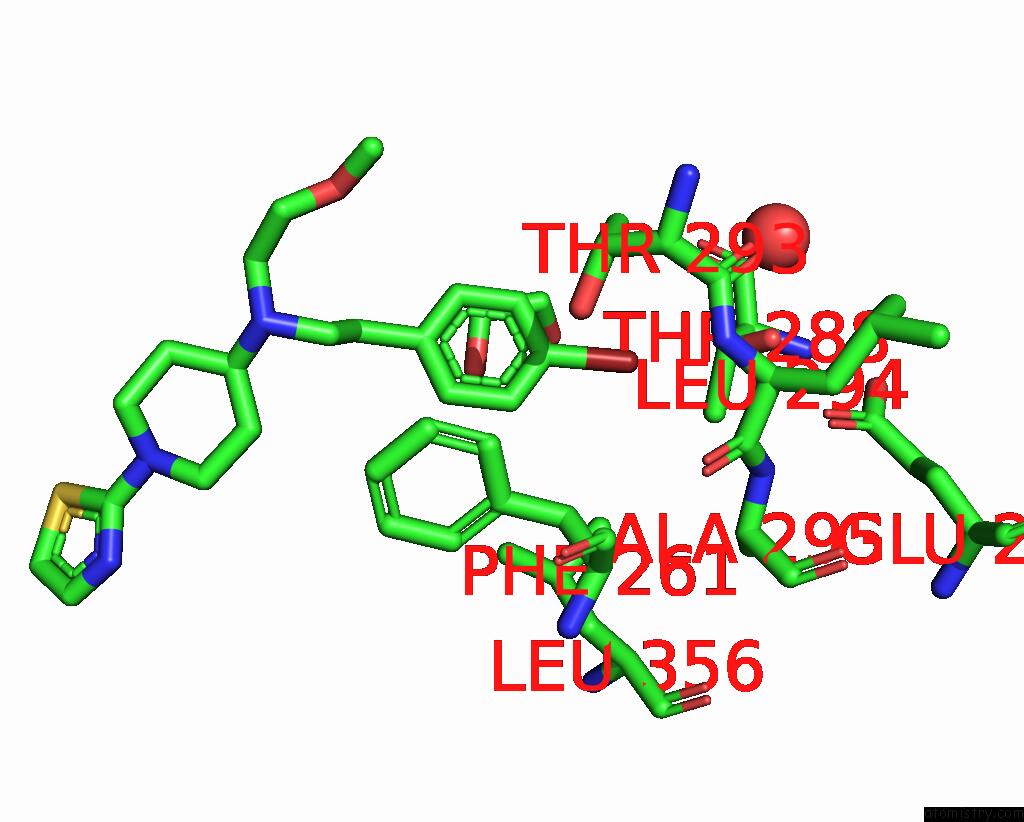
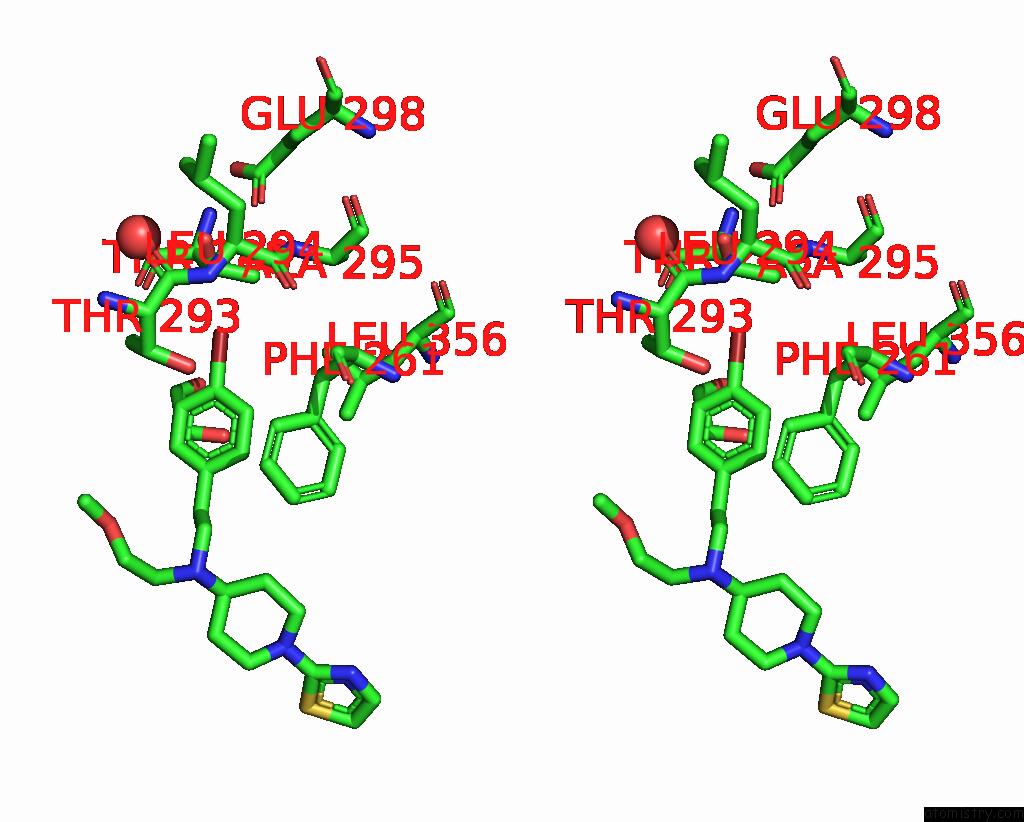
Bromine binding site 4 out of 4 in 8r41
Go back to
Bromine binding site 4 out
of 4 in the Structure of CHI3L1 in Complex with Inhibitor 1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 4 of Structure of CHI3L1 in Complex with Inhibitor 1 within 5.0Å range:
|
Reference:
W.Czestkowski,
L.Krzeminski,
M.C.Piotrowicz,
M.Mazur,
E.Pluta,
G.Andryianau,
R.Koralewski,
K.Matyszewski,
S.Olejniczak,
M.Kowalski,
K.Lisiecka,
R.Koziel,
K.Piwowar,
D.Papiernik,
M.Nowotny,
A.Napiorkowska-Gromadzka,
E.Nowak,
D.Niedzialek,
G.Wieczorek,
A.Siwinska,
T.Rejczak,
K.Jedrzejczak,
K.Mulewski,
J.Olczak,
Z.Zaslona,
A.Golebiowski,
K.Drzewicka,
A.Bartoszewicz.
Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1. J.Med.Chem. 2024.
ISSN: ISSN 0022-2623
PubMed: 38427954
DOI: 10.1021/ACS.JMEDCHEM.3C02255
Page generated: Mon Jul 7 12:35:02 2025
ISSN: ISSN 0022-2623
PubMed: 38427954
DOI: 10.1021/ACS.JMEDCHEM.3C02255
Last articles
Cl in 3N06Cl in 3MZB
Cl in 3MZ9
Cl in 3MZG
Cl in 3MZ0
Cl in 3MZD
Cl in 3MZ1
Cl in 3MYR
Cl in 3MY7
Cl in 3MYX