Bromine »
PDB 2civ-2hb1 »
2fcc »
Bromine in PDB 2fcc: Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site
Enzymatic activity of Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site
All present enzymatic activity of Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site:
3.1.25.1;
3.1.25.1;
Protein crystallography data
The structure of Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site, PDB code: 2fcc
was solved by
G.Golan,
D.O.Zharkov,
A.S.Fernandes,
M.L.Dodson,
A.K.Mccullough,
A.P.Grollman,
R.S.Lloyd,
G.Shoham,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 39.85 / 2.30 |
| Space group | P 64 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 184.059, 184.059, 100.855, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 24.9 / 27.4 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site
(pdb code 2fcc). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 8 binding sites of Bromine where determined in the Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site, PDB code: 2fcc:
Jump to Bromine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Bromine where determined in the Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site, PDB code: 2fcc:
Jump to Bromine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Bromine binding site 1 out of 8 in 2fcc
Go back to
Bromine binding site 1 out
of 8 in the Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site
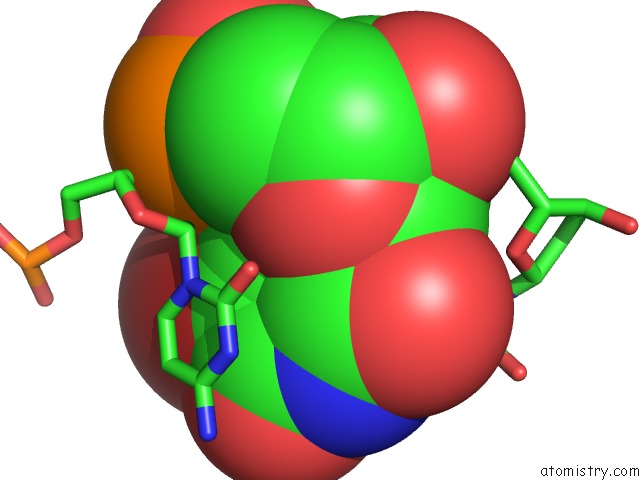
Mono view
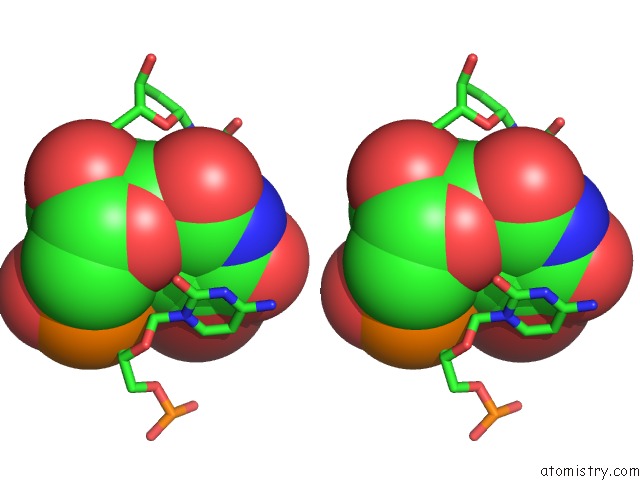
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site within 5.0Å range:
|
Bromine binding site 2 out of 8 in 2fcc
Go back to
Bromine binding site 2 out
of 8 in the Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site
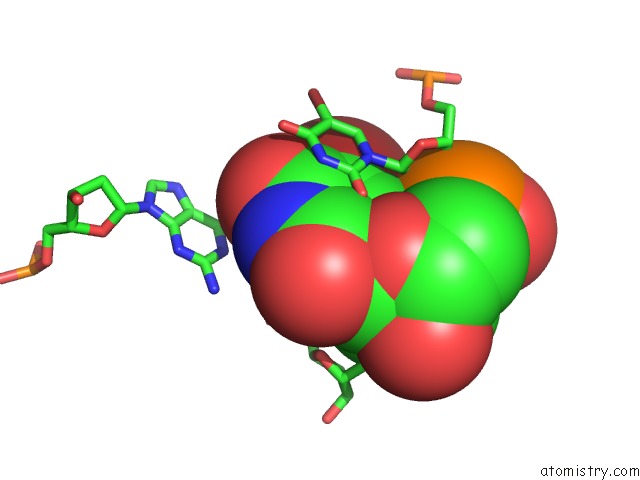
Mono view
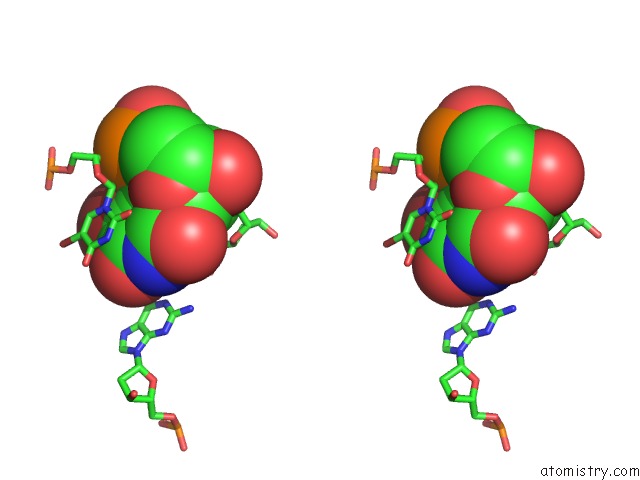
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site within 5.0Å range:
|
Bromine binding site 3 out of 8 in 2fcc
Go back to
Bromine binding site 3 out
of 8 in the Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site
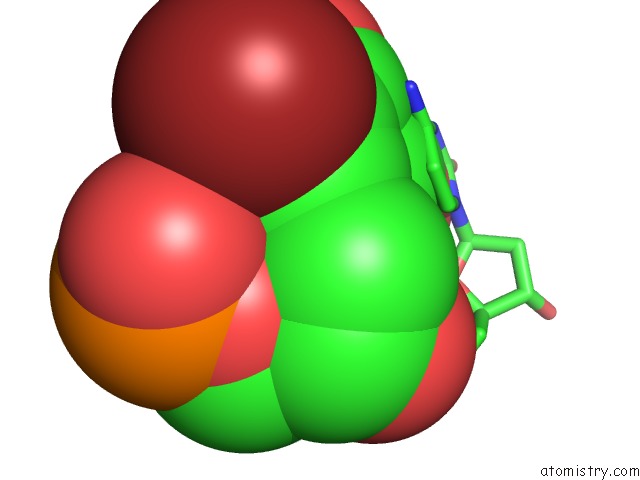
Mono view
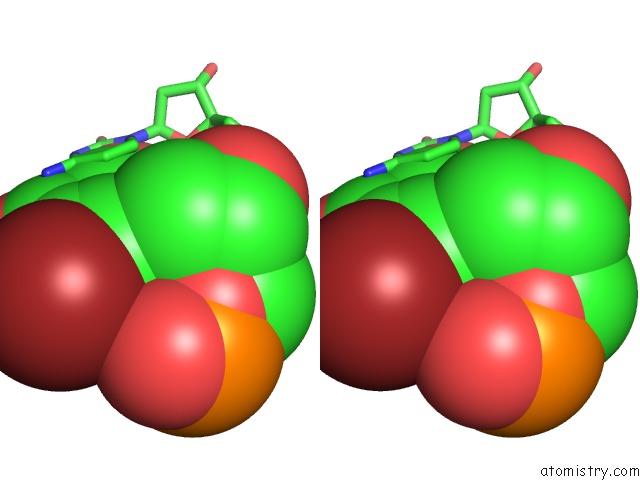
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site within 5.0Å range:
|
Bromine binding site 4 out of 8 in 2fcc
Go back to
Bromine binding site 4 out
of 8 in the Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site
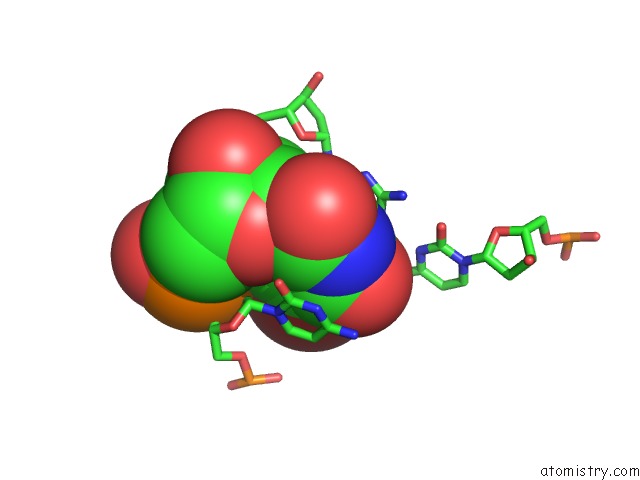
Mono view
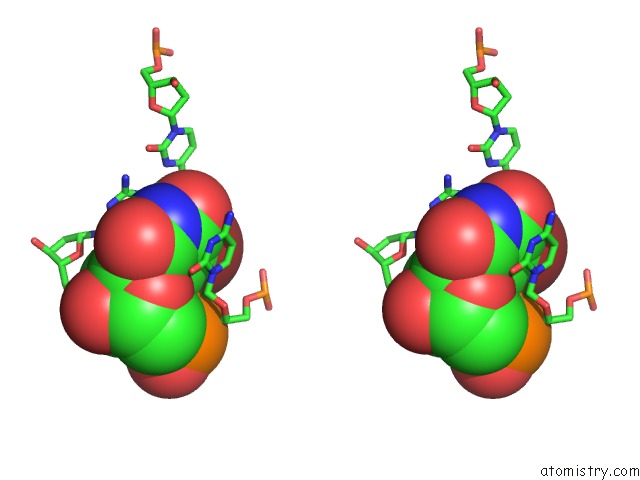
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 4 of Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site within 5.0Å range:
|
Bromine binding site 5 out of 8 in 2fcc
Go back to
Bromine binding site 5 out
of 8 in the Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site
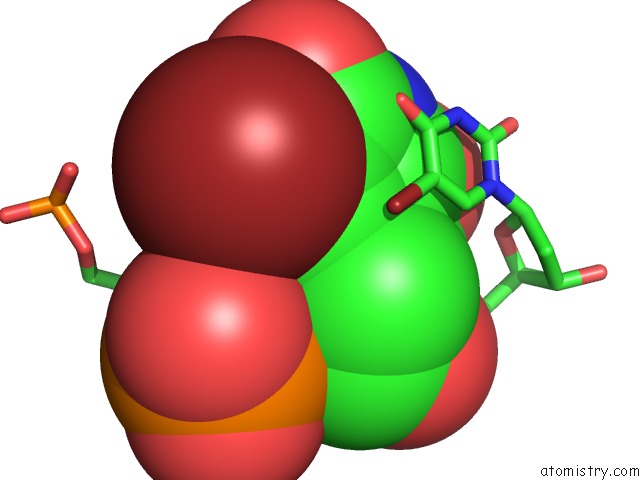
Mono view
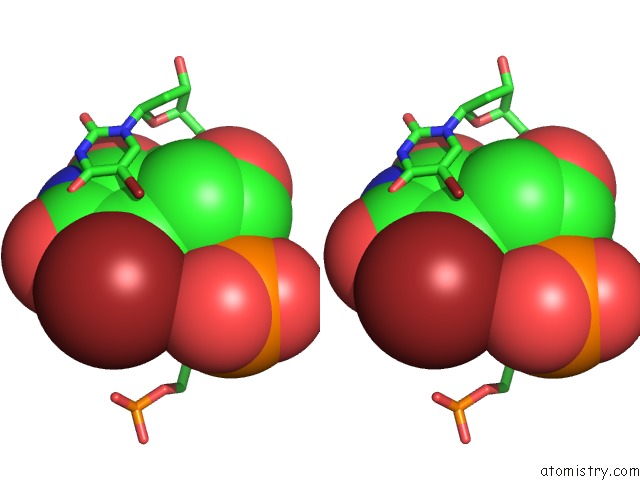
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 5 of Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site within 5.0Å range:
|
Bromine binding site 6 out of 8 in 2fcc
Go back to
Bromine binding site 6 out
of 8 in the Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site
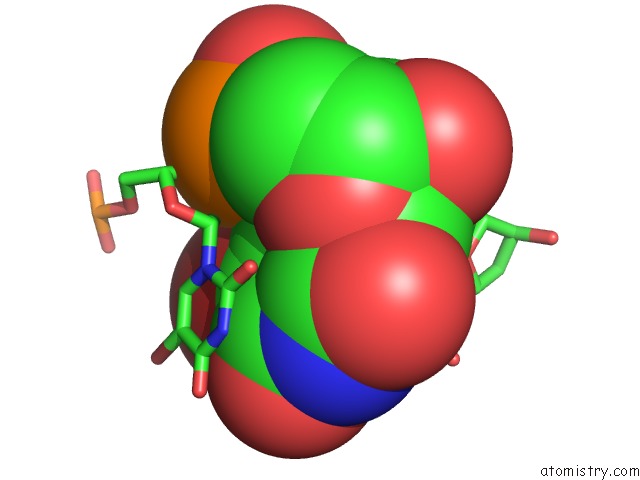
Mono view
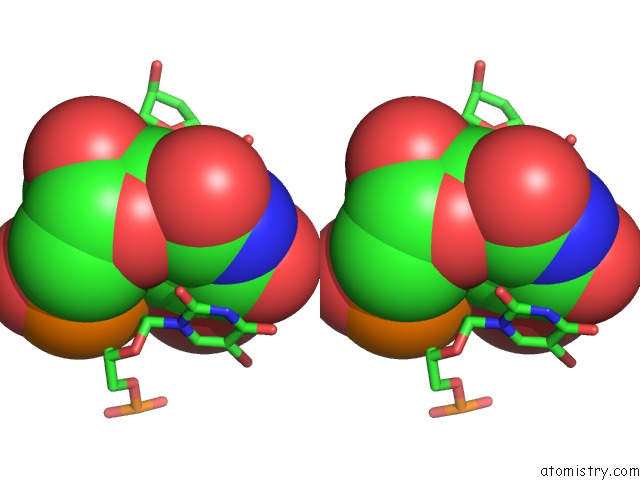
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 6 of Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site within 5.0Å range:
|
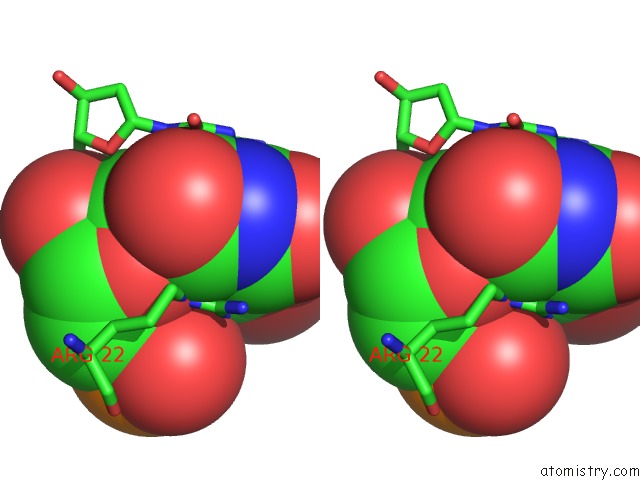
Bromine binding site 7 out of 8 in 2fcc
Go back to
Bromine binding site 7 out
of 8 in the Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site
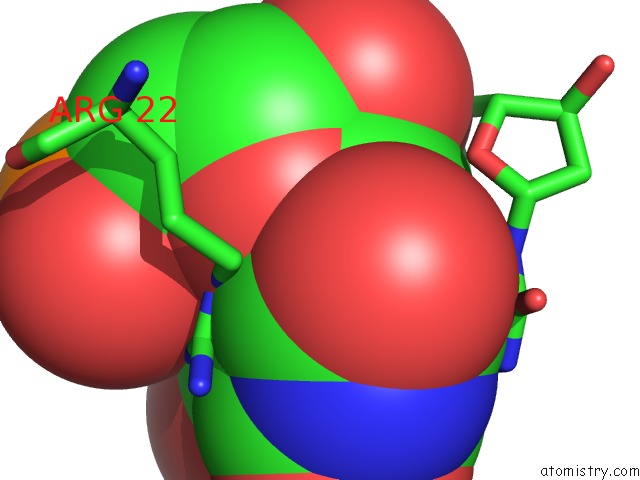
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 7 of Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site within 5.0Å range:
|
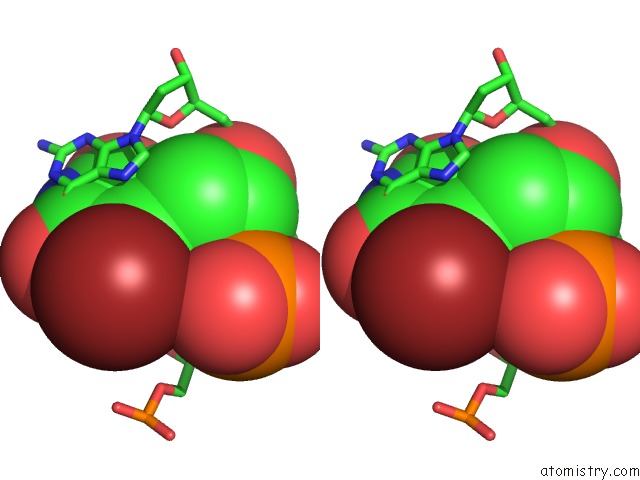
Bromine binding site 8 out of 8 in 2fcc
Go back to
Bromine binding site 8 out
of 8 in the Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site
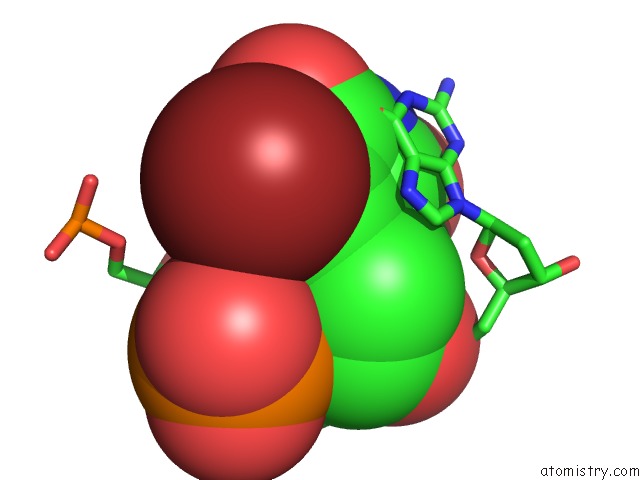
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 8 of Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with A Dna Substrate Containing Abasic Site within 5.0Å range:
|
Reference:
G.Golan,
D.O.Zharkov,
A.P.Grollman,
M.L.Dodson,
A.K.Mccullough,
R.S.Lloyd,
G.Shoham.
Structure of T4 Pyrimidine Dimer Glycosylase in A Reduced Imine Covalent Complex with Abasic Site-Containing Dna. J.Mol.Biol. V. 362 241 2006.
ISSN: ISSN 0022-2836
PubMed: 16916523
DOI: 10.1016/J.JMB.2006.06.059
Page generated: Mon Jul 7 04:13:32 2025
ISSN: ISSN 0022-2836
PubMed: 16916523
DOI: 10.1016/J.JMB.2006.06.059
Last articles
K in 5L9WK in 5L9D
K in 5L88
K in 5KSD
K in 5KSE
K in 5K09
K in 5KOE
K in 5KMT
K in 5KIL
K in 5KIK