Bromine »
PDB 6e4l-6h45 »
6fex »
Bromine in PDB 6fex: DDR1, 2-[4-Bromo-2-Oxo-1'-(1H-Pyrazolo[4,3-B]Pyridine-5-Carbonyl) Spiro[Indole-3,4'-Piperidine]-1-Yl]-N-(2,2,2-Trifluoroethyl) Acetamide, 1.291A, P212121, Rfree=17.4%
Enzymatic activity of DDR1, 2-[4-Bromo-2-Oxo-1'-(1H-Pyrazolo[4,3-B]Pyridine-5-Carbonyl) Spiro[Indole-3,4'-Piperidine]-1-Yl]-N-(2,2,2-Trifluoroethyl) Acetamide, 1.291A, P212121, Rfree=17.4%
All present enzymatic activity of DDR1, 2-[4-Bromo-2-Oxo-1'-(1H-Pyrazolo[4,3-B]Pyridine-5-Carbonyl) Spiro[Indole-3,4'-Piperidine]-1-Yl]-N-(2,2,2-Trifluoroethyl) Acetamide, 1.291A, P212121, Rfree=17.4%:
2.7.10.1;
2.7.10.1;
Protein crystallography data
The structure of DDR1, 2-[4-Bromo-2-Oxo-1'-(1H-Pyrazolo[4,3-B]Pyridine-5-Carbonyl) Spiro[Indole-3,4'-Piperidine]-1-Yl]-N-(2,2,2-Trifluoroethyl) Acetamide, 1.291A, P212121, Rfree=17.4%, PDB code: 6fex
was solved by
M.Stihle,
H.Richter,
J.Benz,
B.Kuhn,
M.G.Rudolph,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 60.47 / 1.29 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 40.834, 61.750, 63.072, 90.00, 106.51, 90.00 |
| R / Rfree (%) | 13.7 / 17.4 |
Other elements in 6fex:
The structure of DDR1, 2-[4-Bromo-2-Oxo-1'-(1H-Pyrazolo[4,3-B]Pyridine-5-Carbonyl) Spiro[Indole-3,4'-Piperidine]-1-Yl]-N-(2,2,2-Trifluoroethyl) Acetamide, 1.291A, P212121, Rfree=17.4% also contains other interesting chemical elements:
| Fluorine | (F) | 3 atoms |
| Iodine | (I) | 4 atoms |
| Chlorine | (Cl) | 4 atoms |
Bromine Binding Sites:
The binding sites of Bromine atom in the DDR1, 2-[4-Bromo-2-Oxo-1'-(1H-Pyrazolo[4,3-B]Pyridine-5-Carbonyl) Spiro[Indole-3,4'-Piperidine]-1-Yl]-N-(2,2,2-Trifluoroethyl) Acetamide, 1.291A, P212121, Rfree=17.4%
(pdb code 6fex). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total only one binding site of Bromine was determined in the DDR1, 2-[4-Bromo-2-Oxo-1'-(1H-Pyrazolo[4,3-B]Pyridine-5-Carbonyl) Spiro[Indole-3,4'-Piperidine]-1-Yl]-N-(2,2,2-Trifluoroethyl) Acetamide, 1.291A, P212121, Rfree=17.4%, PDB code: 6fex:
In total only one binding site of Bromine was determined in the DDR1, 2-[4-Bromo-2-Oxo-1'-(1H-Pyrazolo[4,3-B]Pyridine-5-Carbonyl) Spiro[Indole-3,4'-Piperidine]-1-Yl]-N-(2,2,2-Trifluoroethyl) Acetamide, 1.291A, P212121, Rfree=17.4%, PDB code: 6fex:
Bromine binding site 1 out of 1 in 6fex
Go back to
Bromine binding site 1 out
of 1 in the DDR1, 2-[4-Bromo-2-Oxo-1'-(1H-Pyrazolo[4,3-B]Pyridine-5-Carbonyl) Spiro[Indole-3,4'-Piperidine]-1-Yl]-N-(2,2,2-Trifluoroethyl) Acetamide, 1.291A, P212121, Rfree=17.4%
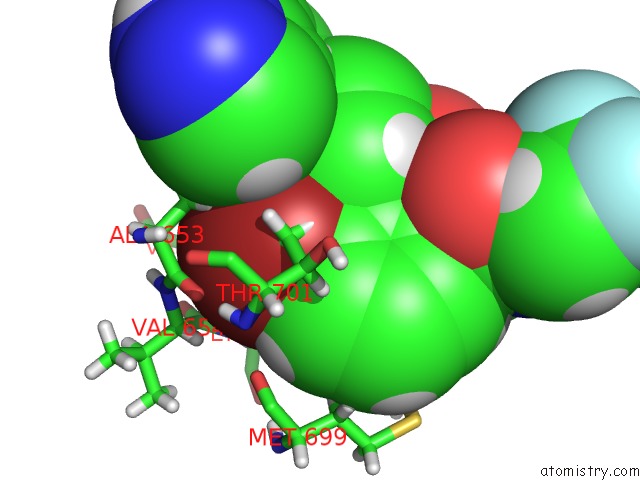
Mono view
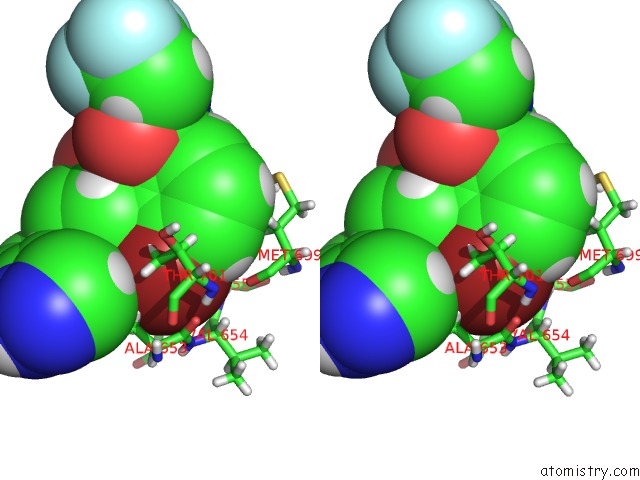
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of DDR1, 2-[4-Bromo-2-Oxo-1'-(1H-Pyrazolo[4,3-B]Pyridine-5-Carbonyl) Spiro[Indole-3,4'-Piperidine]-1-Yl]-N-(2,2,2-Trifluoroethyl) Acetamide, 1.291A, P212121, Rfree=17.4% within 5.0Å range:
|
Reference:
H.Richter,
A.L.Satz,
M.Bedoucha,
B.Buettelmann,
A.C.Petersen,
A.Harmeier,
R.Hermosilla,
R.Hochstrasser,
D.Burger,
B.Gsell,
R.Gasser,
S.Huber,
M.N.Hug,
B.Kocer,
B.Kuhn,
M.Ritter,
M.G.Rudolph,
F.Weibel,
J.Molina-David,
J.J.Kim,
J.V.Santos,
M.Stihle,
G.J.Georges,
R.D.Bonfil,
R.Fridman,
S.Uhles,
S.Moll,
C.Faul,
A.Fornoni,
M.Prunotto.
Dna-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in A Genetic Mouse Model of Alport Syndrome. Acs Chem.Biol. V. 14 37 2019.
ISSN: ESSN 1554-8937
PubMed: 30452219
DOI: 10.1021/ACSCHEMBIO.8B00866
Page generated: Mon Jul 7 09:51:08 2025
ISSN: ESSN 1554-8937
PubMed: 30452219
DOI: 10.1021/ACSCHEMBIO.8B00866
Last articles
K in 6L9ZK in 6LA8
K in 6LAB
K in 6LA9
K in 6KSH
K in 6L5G
K in 6L32
K in 6KY3
K in 6KFL
K in 6KKT