Bromine »
PDB 8adf-8d90 »
8clf »
Bromine in PDB 8clf: Z-SOLQ2BR Bound to Tubulin (T2R-Ttl) Complex
Protein crystallography data
The structure of Z-SOLQ2BR Bound to Tubulin (T2R-Ttl) Complex, PDB code: 8clf
was solved by
M.Wranik,
Q.Bertrand,
M.Kepa,
T.Weinert,
M.Steinmetz,
J.Standfuss,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 15.35 / 2.70 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 106.65, 160.33, 180.95, 90, 90, 90 |
| R / Rfree (%) | 16.8 / 21.7 |
Other elements in 8clf:
The structure of Z-SOLQ2BR Bound to Tubulin (T2R-Ttl) Complex also contains other interesting chemical elements:
| Magnesium | (Mg) | 4 atoms |
| Calcium | (Ca) | 2 atoms |
Bromine Binding Sites:
The binding sites of Bromine atom in the Z-SOLQ2BR Bound to Tubulin (T2R-Ttl) Complex
(pdb code 8clf). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total only one binding site of Bromine was determined in the Z-SOLQ2BR Bound to Tubulin (T2R-Ttl) Complex, PDB code: 8clf:
In total only one binding site of Bromine was determined in the Z-SOLQ2BR Bound to Tubulin (T2R-Ttl) Complex, PDB code: 8clf:
Bromine binding site 1 out of 1 in 8clf
Go back to
Bromine binding site 1 out
of 1 in the Z-SOLQ2BR Bound to Tubulin (T2R-Ttl) Complex
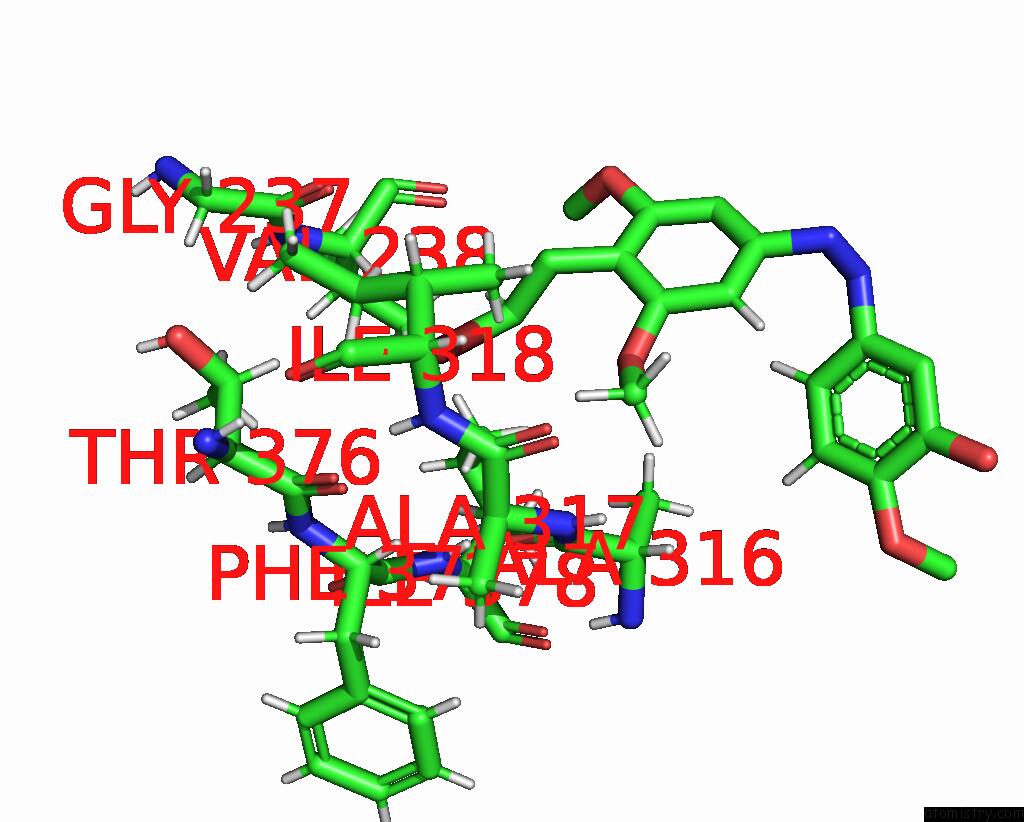
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Z-SOLQ2BR Bound to Tubulin (T2R-Ttl) Complex within 5.0Å range:
|
Reference:
M.Wranik,
M.W.Kepa,
E.V.Beale,
D.James,
Q.Bertrand,
T.Weinert,
A.Furrer,
H.Glover,
D.Gashi,
M.Carrillo,
Y.Kondo,
R.T.Stipp,
G.Khusainov,
K.Nass,
D.Ozerov,
C.Cirelli,
P.J.M.Johnson,
F.Dworkowski,
J.H.Beale,
S.Stubbs,
T.Zamofing,
M.Schneider,
K.Krauskopf,
L.Gao,
O.Thorn-Seshold,
C.Bostedt,
C.Bacellar,
M.O.Steinmetz,
C.Milne,
J.Standfuss.
A Multi-Reservoir Extruder For Time-Resolved Serial Protein Crystallography and Compound Screening at X-Ray Free-Electron Lasers. Nat Commun V. 14 7956 2023.
ISSN: ESSN 2041-1723
PubMed: 38042952
DOI: 10.1038/S41467-023-43523-5
Page generated: Mon Jul 7 12:07:49 2025
ISSN: ESSN 2041-1723
PubMed: 38042952
DOI: 10.1038/S41467-023-43523-5
Last articles
I in 4PNXI in 4PVQ
I in 4PVG
I in 4PGC
I in 4PNS
I in 4P4Z
I in 4P1E
I in 4P4X
I in 4P4Y
I in 4P4W