Bromine »
PDB 1iha-1m9r »
1m67 »
Bromine in PDB 1m67: Crystal Structure of Leishmania Mexicana Gpdh Complexed with Inhibitor 2-Bromo-6-Hydroxy-Purine
Enzymatic activity of Crystal Structure of Leishmania Mexicana Gpdh Complexed with Inhibitor 2-Bromo-6-Hydroxy-Purine
All present enzymatic activity of Crystal Structure of Leishmania Mexicana Gpdh Complexed with Inhibitor 2-Bromo-6-Hydroxy-Purine:
1.1.1.8;
1.1.1.8;
Protein crystallography data
The structure of Crystal Structure of Leishmania Mexicana Gpdh Complexed with Inhibitor 2-Bromo-6-Hydroxy-Purine, PDB code: 1m67
was solved by
J.Choe,
S.Suresh,
G.Wisedchaisri,
K.J.Kennedy,
M.H.Gelb,
W.G.J.Hol,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.50 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 70.503, 70.503, 211.738, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 24.4 / 28.6 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Crystal Structure of Leishmania Mexicana Gpdh Complexed with Inhibitor 2-Bromo-6-Hydroxy-Purine
(pdb code 1m67). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total only one binding site of Bromine was determined in the Crystal Structure of Leishmania Mexicana Gpdh Complexed with Inhibitor 2-Bromo-6-Hydroxy-Purine, PDB code: 1m67:
In total only one binding site of Bromine was determined in the Crystal Structure of Leishmania Mexicana Gpdh Complexed with Inhibitor 2-Bromo-6-Hydroxy-Purine, PDB code: 1m67:
Bromine binding site 1 out of 1 in 1m67
Go back to
Bromine binding site 1 out
of 1 in the Crystal Structure of Leishmania Mexicana Gpdh Complexed with Inhibitor 2-Bromo-6-Hydroxy-Purine
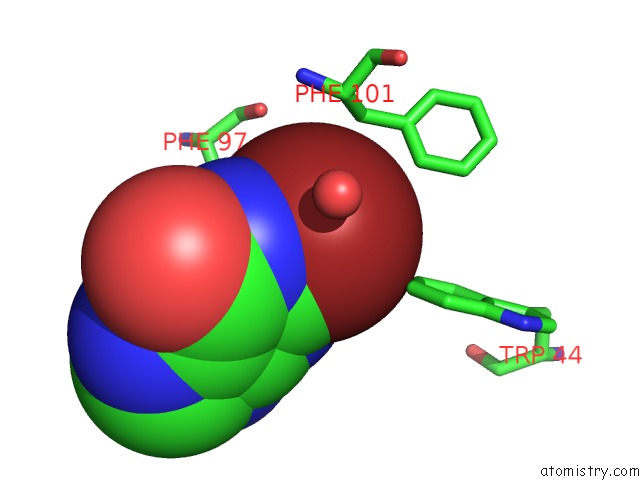
Mono view
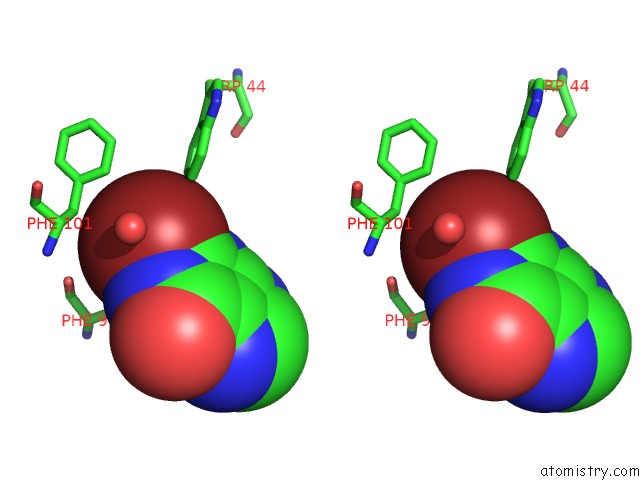
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Crystal Structure of Leishmania Mexicana Gpdh Complexed with Inhibitor 2-Bromo-6-Hydroxy-Purine within 5.0Å range:
|
Reference:
J.Choe,
S.Suresh,
G.Wisedchaisri,
K.J.Kennedy,
M.H.Gelb,
W.G.J.Hol.
Anomalous Differences of Light Elements in Determining Precise Binding Modes of Ligands to Glycerol-3-Phosphate Dehydrogenase Chem.Biol. V. 9 1189 2002.
ISSN: ISSN 1074-5521
PubMed: 12445769
DOI: 10.1016/S1074-5521(02)00243-0
Page generated: Mon Jul 7 03:26:23 2025
ISSN: ISSN 1074-5521
PubMed: 12445769
DOI: 10.1016/S1074-5521(02)00243-0
Last articles
Br in 6MWLBr in 6MWH
Br in 6MEI
Br in 6M97
Br in 6MUG
Br in 6MU8
Br in 6MN8
Br in 6MDA
Br in 6M4T
Br in 6M5B