Bromine »
PDB 2xpn-3bnq »
3aaq »
Bromine in PDB 3aaq: Crystal Structure of LP1NTPDASE From Legionella Pneumophila in Complex with the Inhibitor Arl 67156
Protein crystallography data
The structure of Crystal Structure of LP1NTPDASE From Legionella Pneumophila in Complex with the Inhibitor Arl 67156, PDB code: 3aaq
was solved by
J.P.Vivian,
T.Beddoe,
J.Rossjohn,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 34.44 / 2.00 |
| Space group | P 3 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 101.479, 101.479, 74.867, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.5 / 22.4 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Crystal Structure of LP1NTPDASE From Legionella Pneumophila in Complex with the Inhibitor Arl 67156
(pdb code 3aaq). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 2 binding sites of Bromine where determined in the Crystal Structure of LP1NTPDASE From Legionella Pneumophila in Complex with the Inhibitor Arl 67156, PDB code: 3aaq:
Jump to Bromine binding site number: 1; 2;
In total 2 binding sites of Bromine where determined in the Crystal Structure of LP1NTPDASE From Legionella Pneumophila in Complex with the Inhibitor Arl 67156, PDB code: 3aaq:
Jump to Bromine binding site number: 1; 2;
Bromine binding site 1 out of 2 in 3aaq
Go back to
Bromine binding site 1 out
of 2 in the Crystal Structure of LP1NTPDASE From Legionella Pneumophila in Complex with the Inhibitor Arl 67156
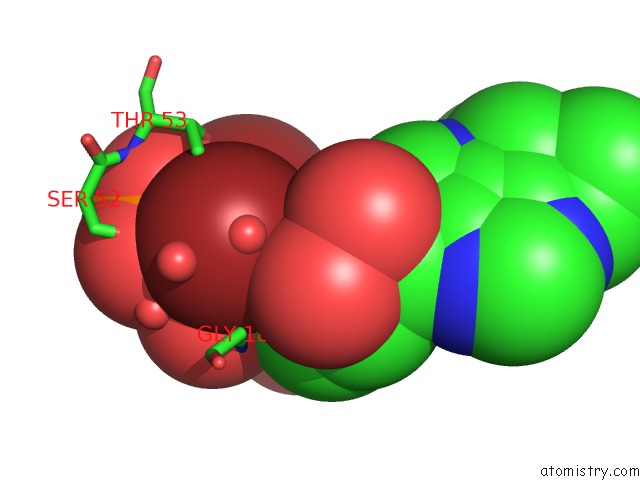
Mono view
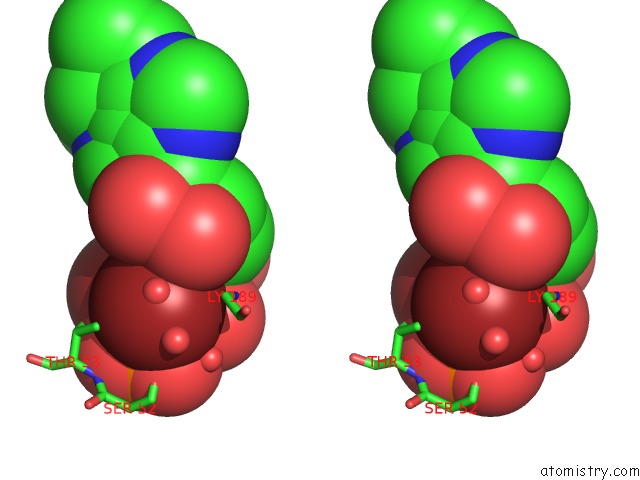
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Crystal Structure of LP1NTPDASE From Legionella Pneumophila in Complex with the Inhibitor Arl 67156 within 5.0Å range:
|
Bromine binding site 2 out of 2 in 3aaq
Go back to
Bromine binding site 2 out
of 2 in the Crystal Structure of LP1NTPDASE From Legionella Pneumophila in Complex with the Inhibitor Arl 67156
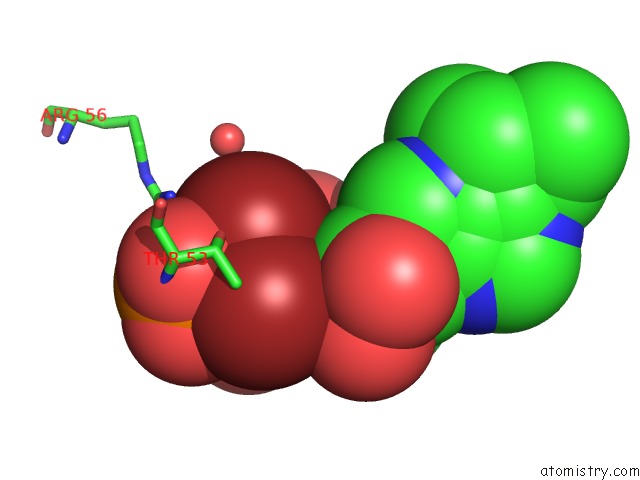
Mono view
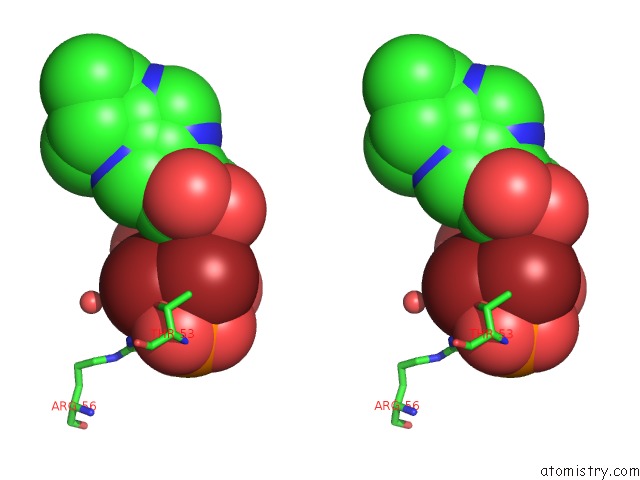
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Crystal Structure of LP1NTPDASE From Legionella Pneumophila in Complex with the Inhibitor Arl 67156 within 5.0Å range:
|
Reference:
J.P.Vivian,
P.Riedmaier,
H.Ge,
J.Le Nours,
F.M.Sansom,
M.C.J.Wilce,
E.Byres,
M.Dias,
J.W.Schmidberger,
P.J.Cowan,
A.J.F.D'apice,
E.L.Hartland,
J.Rossjohn,
T.Beddoe.
Crystal Structure of A Legionella Pneumophila Ecto -Triphosphate Diphosphohydrolase, A Structural and Functional Homolog of the Eukaryotic Ntpdases Structure V. 18 228 2010.
ISSN: ISSN 0969-2126
PubMed: 20159467
DOI: 10.1016/J.STR.2009.11.014
Page generated: Mon Jul 7 05:02:30 2025
ISSN: ISSN 0969-2126
PubMed: 20159467
DOI: 10.1016/J.STR.2009.11.014
Last articles
Br in 5PZTBr in 5PZV
Br in 5OWL
Br in 5POC
Br in 5PH5
Br in 5PO6
Br in 5PBC
Br in 5OWH
Br in 5PA7
Br in 5P9U