Bromine »
PDB 3x41-4alu »
430d »
Bromine in PDB 430d: Structure of Sarcin/Ricin Loop From Rat 28S Rrna
Protein crystallography data
The structure of Structure of Sarcin/Ricin Loop From Rat 28S Rrna, PDB code: 430d
was solved by
C.C.Correll,
A.Munishkin,
Y.L.Chan,
Z.Ren,
I.G.Wool,
T.A.Steitz,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 2.10 |
| Space group | P 61 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 56.830, 56.830, 107.960, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 28 / 33 |
Other elements in 430d:
The structure of Structure of Sarcin/Ricin Loop From Rat 28S Rrna also contains other interesting chemical elements:
| Magnesium | (Mg) | 9 atoms |
Bromine Binding Sites:
The binding sites of Bromine atom in the Structure of Sarcin/Ricin Loop From Rat 28S Rrna
(pdb code 430d). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total only one binding site of Bromine was determined in the Structure of Sarcin/Ricin Loop From Rat 28S Rrna, PDB code: 430d:
In total only one binding site of Bromine was determined in the Structure of Sarcin/Ricin Loop From Rat 28S Rrna, PDB code: 430d:
Bromine binding site 1 out of 1 in 430d
Go back to
Bromine binding site 1 out
of 1 in the Structure of Sarcin/Ricin Loop From Rat 28S Rrna
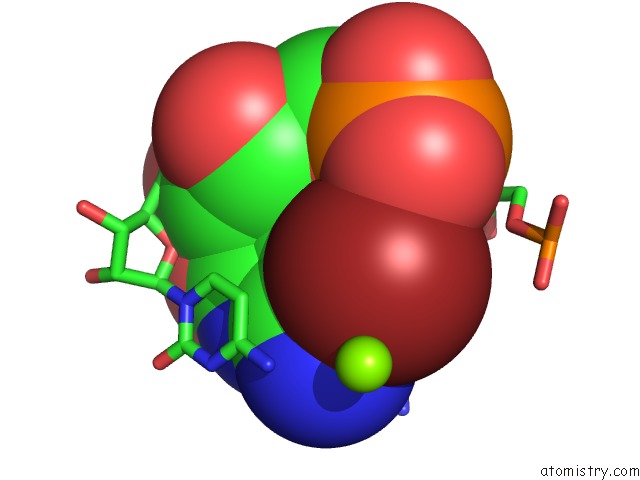
Mono view
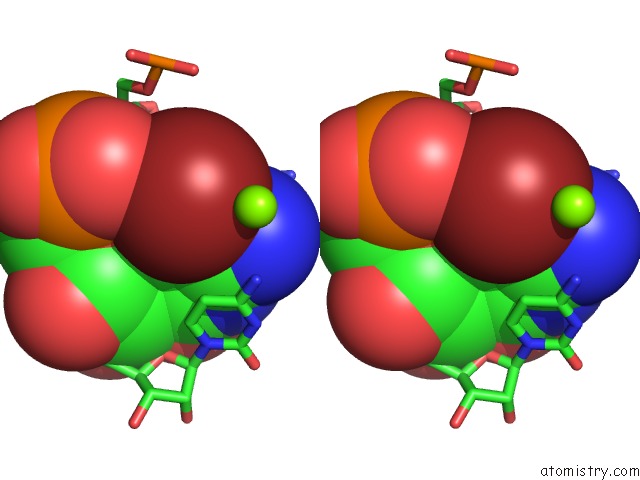
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Structure of Sarcin/Ricin Loop From Rat 28S Rrna within 5.0Å range:
|
Reference:
C.C.Correll,
A.Munishkin,
Y.L.Chan,
Z.Ren,
I.G.Wool,
T.A.Steitz.
Crystal Structure of the Ribosomal Rna Domain Essential For Binding Elongation Factors. Proc.Natl.Acad.Sci.Usa V. 95 13436 1998.
ISSN: ISSN 0027-8424
PubMed: 9811818
DOI: 10.1073/PNAS.95.23.13436
Page generated: Mon Jul 7 06:13:06 2025
ISSN: ISSN 0027-8424
PubMed: 9811818
DOI: 10.1073/PNAS.95.23.13436
Last articles
Cl in 5SFPCl in 5SFG
Cl in 5SF7
Cl in 5SG4
Cl in 5SEZ
Cl in 5SEJ
Cl in 5SEI
Cl in 5SED
Cl in 5SE3
Cl in 5SDS