Bromine »
PDB 4x6i-4z8b »
4yrf »
Bromine in PDB 4yrf: Crystal Structure of T. Cruzi Histidyl-Trna Synthetase in Complex with 5-Bromopyridin-2(1H)-One (Chem 148)
Enzymatic activity of Crystal Structure of T. Cruzi Histidyl-Trna Synthetase in Complex with 5-Bromopyridin-2(1H)-One (Chem 148)
All present enzymatic activity of Crystal Structure of T. Cruzi Histidyl-Trna Synthetase in Complex with 5-Bromopyridin-2(1H)-One (Chem 148):
6.1.1.21;
6.1.1.21;
Protein crystallography data
The structure of Crystal Structure of T. Cruzi Histidyl-Trna Synthetase in Complex with 5-Bromopyridin-2(1H)-One (Chem 148), PDB code: 4yrf
was solved by
C.-Y.Koh,
W.G.J.Hol,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 28.95 / 2.20 |
| Space group | I 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 64.433, 118.814, 65.975, 90.00, 92.39, 90.00 |
| R / Rfree (%) | 21.1 / 26.1 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Crystal Structure of T. Cruzi Histidyl-Trna Synthetase in Complex with 5-Bromopyridin-2(1H)-One (Chem 148)
(pdb code 4yrf). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total only one binding site of Bromine was determined in the Crystal Structure of T. Cruzi Histidyl-Trna Synthetase in Complex with 5-Bromopyridin-2(1H)-One (Chem 148), PDB code: 4yrf:
In total only one binding site of Bromine was determined in the Crystal Structure of T. Cruzi Histidyl-Trna Synthetase in Complex with 5-Bromopyridin-2(1H)-One (Chem 148), PDB code: 4yrf:
Bromine binding site 1 out of 1 in 4yrf
Go back to
Bromine binding site 1 out
of 1 in the Crystal Structure of T. Cruzi Histidyl-Trna Synthetase in Complex with 5-Bromopyridin-2(1H)-One (Chem 148)
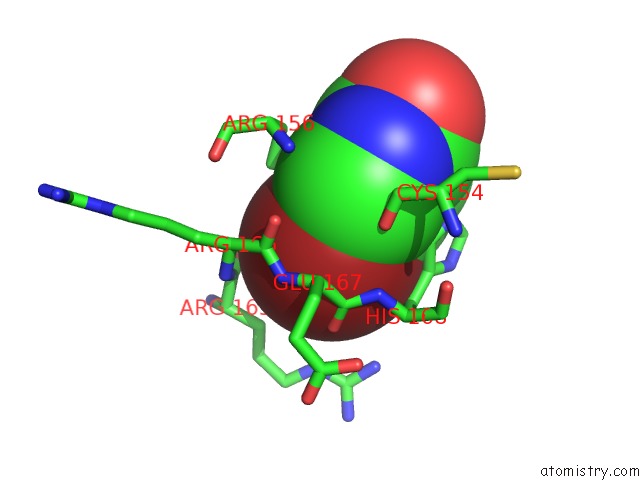
Mono view
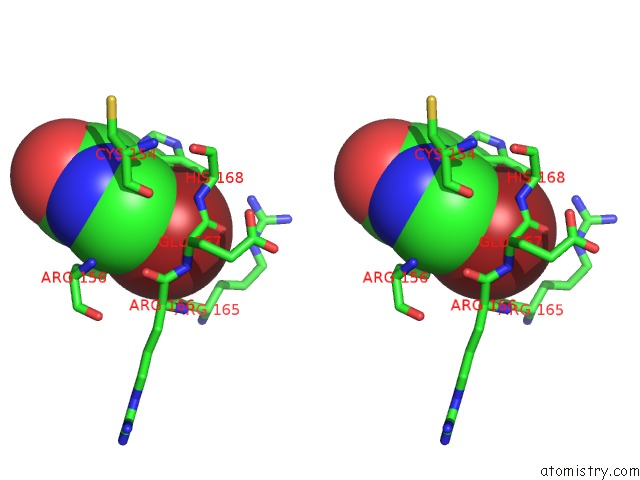
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Crystal Structure of T. Cruzi Histidyl-Trna Synthetase in Complex with 5-Bromopyridin-2(1H)-One (Chem 148) within 5.0Å range:
|
Reference:
C.Y.Koh,
L.Kallur Siddaramaiah,
R.M.Ranade,
J.Nguyen,
T.Jian,
Z.Zhang,
J.R.Gillespie,
F.S.Buckner,
C.L.Verlinde,
E.Fan,
W.G.Hol.
A Binding Hotspot in Trypanosoma Cruzi Histidyl-Trna Synthetase Revealed By Fragment-Based Crystallographic Cocktail Screens. Acta Crystallogr.,Sect.D V. 71 1684 2015.
ISSN: ESSN 1399-0047
PubMed: 26249349
DOI: 10.1107/S1399004715007683
Page generated: Mon Jul 7 07:52:51 2025
ISSN: ESSN 1399-0047
PubMed: 26249349
DOI: 10.1107/S1399004715007683
Last articles
Br in 6YIDBr in 6Y8M
Br in 6Y8I
Br in 6Y00
Br in 6Y02
Br in 6XVG
Br in 6Y3A
Br in 6XQ7
Br in 6XZO
Br in 6XTD