Bromine »
PDB 5i74-5l3j »
5l1g »
Bromine in PDB 5l1g: Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br
Protein crystallography data
The structure of Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br, PDB code: 5l1g
was solved by
M.V.Yelshanskaya,
A.K.Singh,
J.M.Sampson,
A.I.Sobolevsky,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.91 / 4.51 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 92.790, 110.400, 600.140, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 25.7 / 29.5 |
Bromine Binding Sites:
The binding sites of Bromine atom in the Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br
(pdb code 5l1g). This binding sites where shown within
5.0 Angstroms radius around Bromine atom.
In total 5 binding sites of Bromine where determined in the Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br, PDB code: 5l1g:
Jump to Bromine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Bromine where determined in the Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br, PDB code: 5l1g:
Jump to Bromine binding site number: 1; 2; 3; 4; 5;
Bromine binding site 1 out of 5 in 5l1g
Go back to
Bromine binding site 1 out
of 5 in the Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br
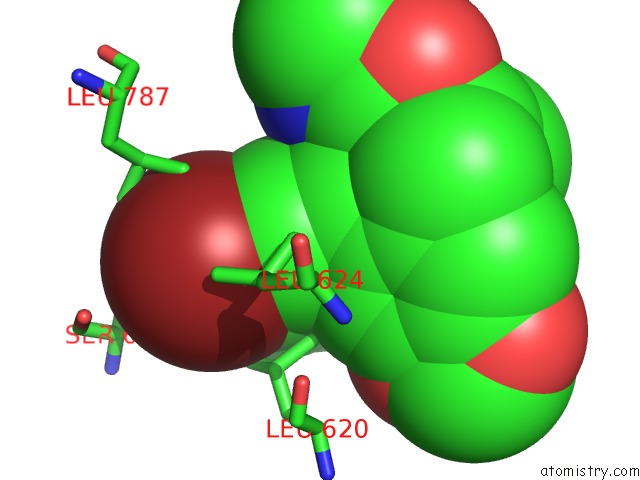
Mono view
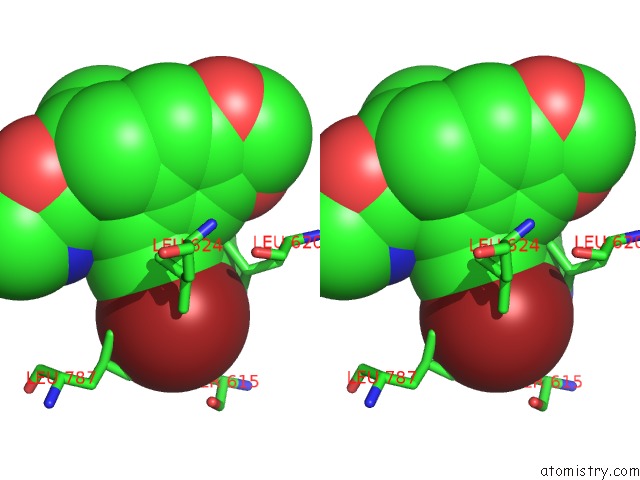
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 1 of Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br within 5.0Å range:
|
Bromine binding site 2 out of 5 in 5l1g
Go back to
Bromine binding site 2 out
of 5 in the Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br
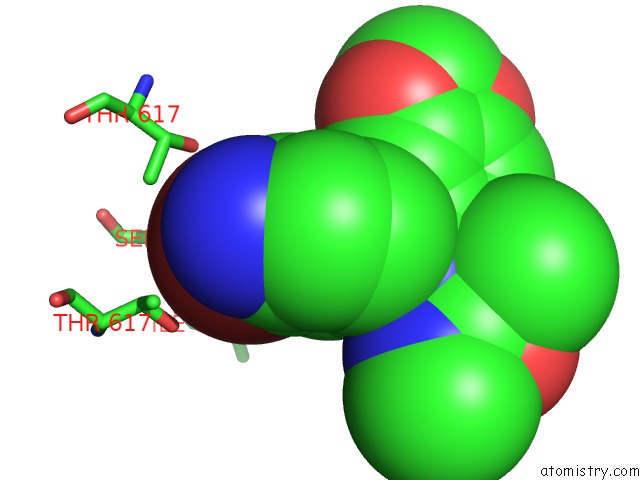
Mono view
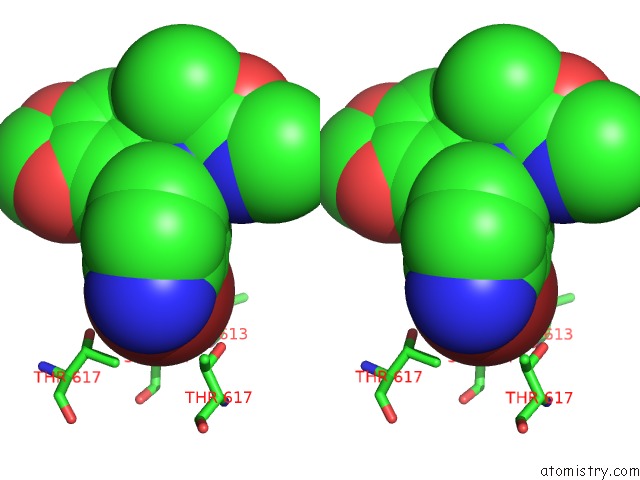
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 2 of Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br within 5.0Å range:
|
Bromine binding site 3 out of 5 in 5l1g
Go back to
Bromine binding site 3 out
of 5 in the Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br
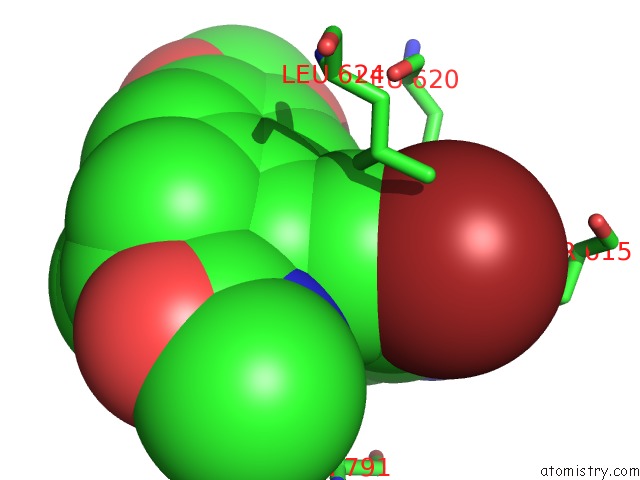
Mono view
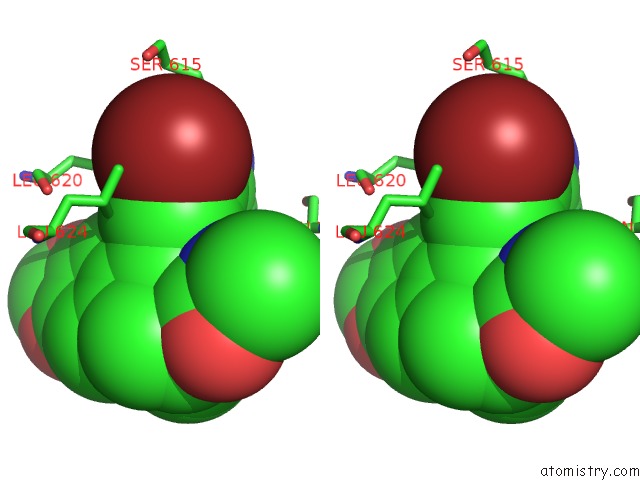
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 3 of Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br within 5.0Å range:
|
Bromine binding site 4 out of 5 in 5l1g
Go back to
Bromine binding site 4 out
of 5 in the Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br
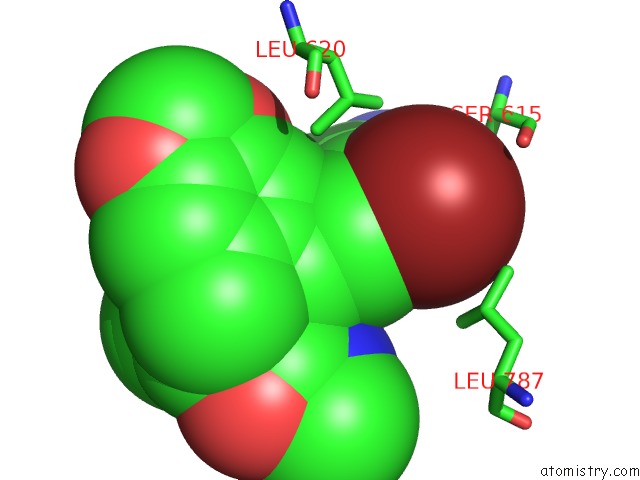
Mono view
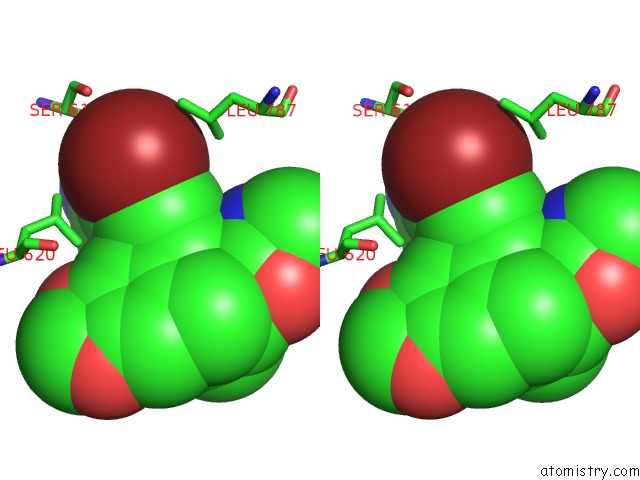
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 4 of Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br within 5.0Å range:
|
Bromine binding site 5 out of 5 in 5l1g
Go back to
Bromine binding site 5 out
of 5 in the Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br
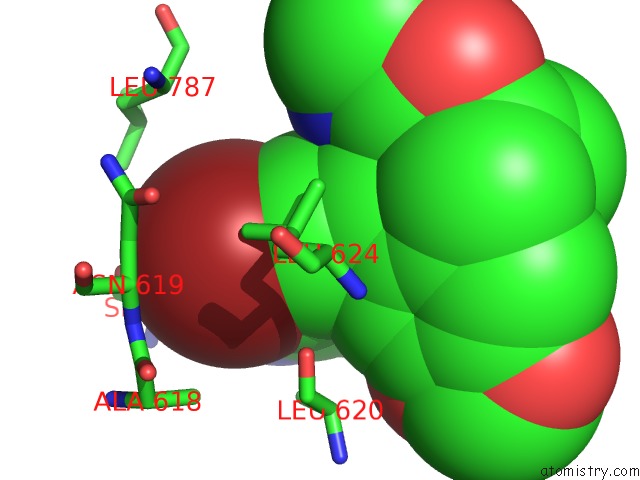
Mono view
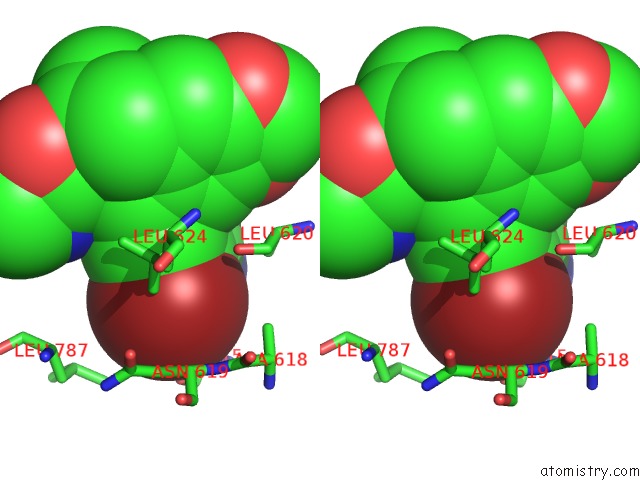
Stereo pair view

Mono view

Stereo pair view
A full contact list of Bromine with other atoms in the Br binding
site number 5 of Ampa Subtype Ionotropic Glutamate Receptor GLUA2 in Complex with Gyki- Br within 5.0Å range:
|
Reference:
M.V.Yelshanskaya,
A.K.Singh,
J.M.Sampson,
C.Narangoda,
M.Kurnikova,
A.I.Sobolevsky.
Structural Bases of Noncompetitive Inhibition of Ampa-Subtype Ionotropic Glutamate Receptors By Antiepileptic Drugs. Neuron V. 91 1305 2016.
ISSN: ISSN 0896-6273
PubMed: 27618672
DOI: 10.1016/J.NEURON.2016.08.012
Page generated: Mon Jul 7 08:33:53 2025
ISSN: ISSN 0896-6273
PubMed: 27618672
DOI: 10.1016/J.NEURON.2016.08.012
Last articles
Br in 6TQGBr in 6SYC
Br in 6TOS
Br in 6TLS
Br in 6TLW
Br in 6TLV
Br in 6TLR
Br in 6TLO
Br in 6TLP
Br in 6TI9